信息传递神经网络 (MPNN) 用于分子属性预测
作者: akensert
创建日期: 2021/08/16
最后修改: 2021/12/27
描述: 实现一个 MPNN 以预测血脑屏障的通透性。
介绍
在本教程中,我们将实现一种称为 _ 信息传递神经网络 _ (MPNN) 的图神经网络 (GNN),以预测图属性。具体来说,我们将实现一个 MPNN 来预测一种名为 _ 血脑屏障通透性 _ (BBBP) 的分子属性。
动机:由于分子自然表示为无向图 G = (V, E),其中 V 是顶点 (节点; 原子) 的集合,E 是边 (键) 的集合,GNN(如 MPNN)被证明是一种有用的方法来预测分子属性。
到目前为止,更传统的方法,如随机森林、支持向量机等,通常被用于预测分子属性。与 GNN 相比,这些传统方法通常在预先计算的分子特征上运行,如分子量、极性、电荷、碳原子数等。虽然这些分子特征证明能够很好地预测各种分子属性,但有人假设在这些更“原始”、“低级”的特征上操作可能会更好。
参考文献
近年来,很多努力被投入到为图数据(包括分子图)开发神经网络。关于图神经网络的总结,参见例如 图神经网络的综合调查 和 图神经网络:方法与应用的综述;关于在本教程中实现的特定图神经网络的进一步阅读可参考 量子化学的神经信息传递 和 DeepChem 的 MPNNModel。
环境设置
安装 RDKit 和其他依赖
(以下文本摘自 这个教程)。
RDKit 是用 C++ 和 Python 编写的化学信息学和机器学习软件的集合。在本教程中,RDKit 被用于便捷高效地将 SMILES 转换为分子对象,并从中获得原子和键的集合。
SMILES 以 ASCII 字符串的形式表达给定分子的结构。SMILES 字符串是一种紧凑编码,对于较小的分子,相对而言比较易于人类阅读。将分子编码为字符串既缓解了数据库和/或网络搜索给定分子的困难,也促进了搜索。RDKit 使用算法准确地将给定的 SMILES 转换为分子对象,然后可以用来计算大量的分子属性/特征。
请注意,RDKit 通常通过 Conda 安装。然而,得益于 rdkit_platform_wheels,RDKit 现在可以(出于本教程的目的)通过 pip 轻松安装,如下所示:
pip -q install rdkit-pypi
为了方便和高效地读取 csv 文件和可视化,下面需要安装:
pip -q install pandas
pip -q install Pillow
pip -q install matplotlib
pip -q install pydot
sudo apt-get -qq install graphviz
导入包
import os
# 临时抑制 tf 日志
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import warnings
from rdkit import Chem
from rdkit import RDLogger
from rdkit.Chem.Draw import IPythonConsole
from rdkit.Chem.Draw import MolsToGridImage
# 临时抑制警告和 RDKit 日志
warnings.filterwarnings("ignore")
RDLogger.DisableLog("rdApp.*")
np.random.seed(42)
tf.random.set_seed(42)
数据集
有关数据集的信息可以在 一种针对计算血脑屏障穿透建模的贝叶斯方法 和 MoleculeNet:分子机器学习的基准 中找到。数据集将从 MoleculeNet.org 下载。
关于
数据集包含 2,050 个分子。每个分子都有 名称、标签 和 SMILES 字符串。
血脑屏障(BBB)是一个隔离血液与脑外细胞液的膜,因此阻止大多数药物(分子)到达大脑。因此, BBBP 对于开发旨在靶向中枢神经系统的新药物的研究至关重要。此数据集的标签是二元的(1 或 0),指示分子的通透性。
csv_path = keras.utils.get_file(
"BBBP.csv", "https://deepchemdata.s3-us-west-1.amazonaws.com/datasets/BBBP.csv"
)
df = pd.read_csv(csv_path, usecols=[1, 2, 3])
df.iloc[96:104]
| 名称 | p_np | smiles | |
|---|---|---|---|
| 96 | cefoxitin | 1 | CO[C@]1(NC(=O)Cc2sccc2)[C@H]3SCC(=C(N3C1=O)C(O... |
| 97 | Org34167 | 1 | NC(CC=C)c1ccccc1c2noc3c2cccc3 |
| 98 | 9-OH Risperidone | 1 | OC1C(N2CCC1)=NC(C)=C(CCN3CCC(CC3)c4c5ccc(F)cc5... |
| 99 | 对乙酰氨基酚 | 1 | CC(=O)Nc1ccc(O)cc1 |
| 100 | 乙酰水杨酸盐 | 0 | CC(=O)Oc1ccccc1C(O)=O |
| 101 | 别嘌醇 | 0 | O=C1N=CN=C2NNC=C12 |
| 102 | 阿莫斯达 | 0 | CCCCC[C@H](O)/C=C/[C@H]1[C@H](O)CC(=O)[C@@H]1C... |
| 103 | 氨茶碱 | 0 | CN1C(=O)N(C)c2nc[nH]c2C1=O.CN3C(=O)N(C)c4nc[nH... |
定义特征
为了编码原子和键的特征(稍后我们将需要这些特征),我们将分别定义两个类:AtomFeaturizer 和 BondFeaturizer。
为了减少代码行数,即保持本教程简短而简洁,仅考虑一 handful 的(原子和键)特征:[原子特征] 符号(元素)、 价电子数、 氢键数、 轨道杂化、 [键特征] (共价) 键类型 和 共轭。
class Featurizer:
def __init__(self, allowable_sets):
self.dim = 0
self.features_mapping = {}
for k, s in allowable_sets.items():
s = sorted(list(s))
self.features_mapping[k] = dict(zip(s, range(self.dim, len(s) + self.dim)))
self.dim += len(s)
def encode(self, inputs):
output = np.zeros((self.dim,))
for name_feature, feature_mapping in self.features_mapping.items():
feature = getattr(self, name_feature)(inputs)
if feature not in feature_mapping:
continue
output[feature_mapping[feature]] = 1.0
return output
class AtomFeaturizer(Featurizer):
def __init__(self, allowable_sets):
super().__init__(allowable_sets)
def symbol(self, atom):
return atom.GetSymbol()
def n_valence(self, atom):
return atom.GetTotalValence()
def n_hydrogens(self, atom):
return atom.GetTotalNumHs()
def hybridization(self, atom):
return atom.GetHybridization().name.lower()
class BondFeaturizer(Featurizer):
def __init__(self, allowable_sets):
super().__init__(allowable_sets)
self.dim += 1
def encode(self, bond):
output = np.zeros((self.dim,))
if bond is None:
output[-1] = 1.0
return output
output = super().encode(bond)
return output
def bond_type(self, bond):
return bond.GetBondType().name.lower()
def conjugated(self, bond):
return bond.GetIsConjugated()
atom_featurizer = AtomFeaturizer(
allowable_sets={
"symbol": {"B", "Br", "C", "Ca", "Cl", "F", "H", "I", "N", "Na", "O", "P", "S"},
"n_valence": {0, 1, 2, 3, 4, 5, 6},
"n_hydrogens": {0, 1, 2, 3, 4},
"hybridization": {"s", "sp", "sp2", "sp3"},
}
)
bond_featurizer = BondFeaturizer(
allowable_sets={
"bond_type": {"single", "double", "triple", "aromatic"},
"conjugated": {True, False},
}
)
生成图形
在我们能够从SMILES生成完整图形之前,我们需要实现以下功能:
-
molecule_from_smiles,该函数接受一个SMILES作为输入并返回一个分子对象。这一切由RDKit处理。 -
graph_from_molecule,该函数接受一个分子对象作为输入并返回一个图形,表示为三元组(atom_features, bond_features, pair_indices)。为此,我们将利用之前定义的类。
最后,我们现在可以实现函数graphs_from_smiles,该函数在训练、验证和测试数据集的所有SMILES上应用(1)功能,随后(2)。
注意:尽管建议对该数据集进行骨架拆分(见这里),为了简单起见,进行了简单的随机拆分。
def molecule_from_smiles(smiles):
# MolFromSmiles(m, sanitize=True) 应当等同于
# MolFromSmiles(m, sanitize=False) -> SanitizeMol(m) -> AssignStereochemistry(m, ...)
molecule = Chem.MolFromSmiles(smiles, sanitize=False)
# 如果消毒失败,则捕获错误,并尝试再次执行
# 导致错误的消毒步骤
flag = Chem.SanitizeMol(molecule, catchErrors=True)
if flag != Chem.SanitizeFlags.SANITIZE_NONE:
Chem.SanitizeMol(molecule, sanitizeOps=Chem.SanitizeFlags.SANITIZE_ALL ^ flag)
Chem.AssignStereochemistry(molecule, cleanIt=True, force=True)
return molecule
def graph_from_molecule(molecule):
# 初始化图形
atom_features = []
bond_features = []
pair_indices = []
for atom in molecule.GetAtoms():
atom_features.append(atom_featurizer.encode(atom))
# 添加自循环
pair_indices.append([atom.GetIdx(), atom.GetIdx()])
bond_features.append(bond_featurizer.encode(None))
for neighbor in atom.GetNeighbors():
bond = molecule.GetBondBetweenAtoms(atom.GetIdx(), neighbor.GetIdx())
pair_indices.append([atom.GetIdx(), neighbor.GetIdx()])
bond_features.append(bond_featurizer.encode(bond))
return np.array(atom_features), np.array(bond_features), np.array(pair_indices)
def graphs_from_smiles(smiles_list):
# 初始化图形
atom_features_list = []
bond_features_list = []
pair_indices_list = []
for smiles in smiles_list:
molecule = molecule_from_smiles(smiles)
atom_features, bond_features, pair_indices = graph_from_molecule(molecule)
atom_features_list.append(atom_features)
bond_features_list.append(bond_features)
pair_indices_list.append(pair_indices)
# 将列表转换为不规则张量,以便后续的tf.data.Dataset
return (
tf.ragged.constant(atom_features_list, dtype=tf.float32),
tf.ragged.constant(bond_features_list, dtype=tf.float32),
tf.ragged.constant(pair_indices_list, dtype=tf.int64),
)
# 打乱从0到2049的索引数组
permuted_indices = np.random.permutation(np.arange(df.shape[0]))
# 训练集:80 % 的数据
train_index = permuted_indices[: int(df.shape[0] * 0.8)]
x_train = graphs_from_smiles(df.iloc[train_index].smiles)
y_train = df.iloc[train_index].p_np
# 验证集:19 % 的数据
valid_index = permuted_indices[int(df.shape[0] * 0.8) : int(df.shape[0] * 0.99)]
x_valid = graphs_from_smiles(df.iloc[valid_index].smiles)
y_valid = df.iloc[valid_index].p_np
# 测试集:1 % 的数据
test_index = permuted_indices[int(df.shape[0] * 0.99) :]
x_test = graphs_from_smiles(df.iloc[test_index].smiles)
y_test = df.iloc[test_index].p_np
测试函数
print(f"Name:\t{df.name[100]}\nSMILES:\t{df.smiles[100]}\nBBBP:\t{df.p_np[100]}")
molecule = molecule_from_smiles(df.iloc[100].smiles)
print("Molecule:")
molecule
Name:acetylsalicylate
SMILES:CC(=O)Oc1ccccc1C(O)=O
BBBP:0
Molecule:

graph = graph_from_molecule(molecule)
print("Graph (including self-loops):")
print("\tatom features\t", graph[0].shape)
print("\tbond features\t", graph[1].shape)
print("\tpair indices\t", graph[2].shape)
Graph (including self-loops):
atom features (13, 29)
bond features (39, 7)
pair indices (39, 2)
创建 tf.data.Dataset
在本教程中,MPNN实现将作为输入(每次迭代)一个单一图形。因此,对于一批(子)图(分子),我们需要将它们合并为一个单一图形(我们将此图称为全局图)。这个全局图是一个不连通的图形,其中每个子图彼此完全独立。
def prepare_batch(x_batch, y_batch):
"""将批次的(子)图合并为一个单一的全局(不相连)图
"""
atom_features, bond_features, pair_indices = x_batch
# 获取每个图(分子)的原子和键的数量
num_atoms = atom_features.row_lengths()
num_bonds = bond_features.row_lengths()
# 获取分区索引(molecule_indicator),稍后将在模型中用于
# 从全局图收集(子)图
molecule_indices = tf.range(len(num_atoms))
molecule_indicator = tf.repeat(molecule_indices, num_atoms)
# 将(子)图合并为一个全局(不相连)图。将“增量”添加到
# “pair_indices”(并合并不规则张量)实现全局图
gather_indices = tf.repeat(molecule_indices[:-1], num_bonds[1:])
increment = tf.cumsum(num_atoms[:-1])
increment = tf.pad(tf.gather(increment, gather_indices), [(num_bonds[0], 0)])
pair_indices = pair_indices.merge_dims(outer_axis=0, inner_axis=1).to_tensor()
pair_indices = pair_indices + increment[:, tf.newaxis]
atom_features = atom_features.merge_dims(outer_axis=0, inner_axis=1).to_tensor()
bond_features = bond_features.merge_dims(outer_axis=0, inner_axis=1).to_tensor()
return (atom_features, bond_features, pair_indices, molecule_indicator), y_batch
def MPNNDataset(X, y, batch_size=32, shuffle=False):
dataset = tf.data.Dataset.from_tensor_slices((X, (y)))
if shuffle:
dataset = dataset.shuffle(1024)
return dataset.batch(batch_size).map(prepare_batch, -1).prefetch(-1)
模型
MPNN模型可以采用各种形状和形式。在本教程中,我们将实现一个基于原始论文 用于量子化学的神经消息传递和 DeepChem的MPNNModel的MPNN。 本教程的MPNN由三个阶段组成:消息传递、读取和分类。
消息传递
消息传递步骤本身由两个部分组成:
-
边缘网络,它基于它们之间的边缘特征(
e_{vw_{i}}),将来自v的w_{i}的消息传递给v,从而得出更新的节点(状态)v'。w_{i}表示v的第i个邻居。 -
门控递归单元 (GRU),它以最新的节点状态为输入,并根据先前的节点状态进行更新。换句话说,最新的节点状态作为GRU的输入,而先前的节点状态则包含在GRU的记忆状态中。这允许信息从一个节点状态(例如
v)流向另一个(例如v'')。
重要的是,步骤(1)和(2)会重复进行k步,在每一步1...k中,从v聚合的信息的半径(或跳数)增加1。
class EdgeNetwork(layers.Layer):
def build(self, input_shape):
self.atom_dim = input_shape[0][-1]
self.bond_dim = input_shape[1][-1]
self.kernel = self.add_weight(
shape=(self.bond_dim, self.atom_dim * self.atom_dim),
initializer="glorot_uniform",
name="kernel",
)
self.bias = self.add_weight(
shape=(self.atom_dim * self.atom_dim), initializer="zeros", name="bias",
)
self.built = True
def call(self, inputs):
atom_features, bond_features, pair_indices = inputs
# 对边特征应用线性变换
bond_features = tf.matmul(bond_features, self.kernel) + self.bias
# 为后续邻域聚合进行重塑
bond_features = tf.reshape(bond_features, (-1, self.atom_dim, self.atom_dim))
# 获取邻居的原子特征
atom_features_neighbors = tf.gather(atom_features, pair_indices[:, 1])
atom_features_neighbors = tf.expand_dims(atom_features_neighbors, axis=-1)
# 应用邻域聚合
transformed_features = tf.matmul(bond_features, atom_features_neighbors)
transformed_features = tf.squeeze(transformed_features, axis=-1)
aggregated_features = tf.math.unsorted_segment_sum(
transformed_features,
pair_indices[:, 0],
num_segments=tf.shape(atom_features)[0],
)
return aggregated_features
class MessagePassing(layers.Layer):
def __init__(self, units, steps=4, **kwargs):
super().__init__(**kwargs)
self.units = units
self.steps = steps
def build(self, input_shape):
self.atom_dim = input_shape[0][-1]
self.message_step = EdgeNetwork()
self.pad_length = max(0, self.units - self.atom_dim)
self.update_step = layers.GRUCell(self.atom_dim + self.pad_length)
self.built = True
def call(self, inputs):
atom_features, bond_features, pair_indices = inputs
# 如果所需单元数超过原子特征维度,则填充原子特征。
# 或者,这里可以使用一个全连接层。
atom_features_updated = tf.pad(atom_features, [(0, 0), (0, self.pad_length)])
# 进行多步消息传递
for i in range(self.steps):
# 从邻居处聚合信息
atom_features_aggregated = self.message_step(
[atom_features_updated, bond_features, pair_indices]
)
# 通过一轮GRU更新节点状态
atom_features_updated, _ = self.update_step(
atom_features_aggregated, atom_features_updated
)
return atom_features_updated
读取
当消息传递过程结束时,k步聚合的节点状态将被划分到子图中(对应于批次中的每个分子),并随后减少为图级嵌入。在 原始论文中,使用了 集到集层用于此目的。 然而,在本教程中,将使用转换器编码器 + 平均池化。具体来说:
- k步聚合的节点状态将被划分为子图(对应于批次中的每个分子);
- 每个子图将被填充以匹配节点数最多的子图,然后进行
tf.stack(...); - 编码子图的(堆叠填充)张量(每个子图包含一组节点状态)将被掩蔽,以确保填充不会干扰训练;
- 最后,张量被传递给变压器,然后进行平均池化。
class PartitionPadding(layers.Layer):
def __init__(self, batch_size, **kwargs):
super().__init__(**kwargs)
self.batch_size = batch_size
def call(self, inputs):
atom_features, molecule_indicator = inputs
# 获取子图
atom_features_partitioned = tf.dynamic_partition(
atom_features, molecule_indicator, self.batch_size
)
# 填充和堆叠子图
num_atoms = [tf.shape(f)[0] for f in atom_features_partitioned]
max_num_atoms = tf.reduce_max(num_atoms)
atom_features_stacked = tf.stack(
[
tf.pad(f, [(0, max_num_atoms - n), (0, 0)])
for f, n in zip(atom_features_partitioned, num_atoms)
],
axis=0,
)
# 移除空子图(通常是数据集中的最后一批)
gather_indices = tf.where(tf.reduce_sum(atom_features_stacked, (1, 2)) != 0)
gather_indices = tf.squeeze(gather_indices, axis=-1)
return tf.gather(atom_features_stacked, gather_indices, axis=0)
class TransformerEncoderReadout(layers.Layer):
def __init__(
self, num_heads=8, embed_dim=64, dense_dim=512, batch_size=32, **kwargs
):
super().__init__(**kwargs)
self.partition_padding = PartitionPadding(batch_size)
self.attention = layers.MultiHeadAttention(num_heads, embed_dim)
self.dense_proj = keras.Sequential(
[layers.Dense(dense_dim, activation="relu"), layers.Dense(embed_dim),]
)
self.layernorm_1 = layers.LayerNormalization()
self.layernorm_2 = layers.LayerNormalization()
self.average_pooling = layers.GlobalAveragePooling1D()
def call(self, inputs):
x = self.partition_padding(inputs)
padding_mask = tf.reduce_any(tf.not_equal(x, 0.0), axis=-1)
padding_mask = padding_mask[:, tf.newaxis, tf.newaxis, :]
attention_output = self.attention(x, x, attention_mask=padding_mask)
proj_input = self.layernorm_1(x + attention_output)
proj_output = self.layernorm_2(proj_input + self.dense_proj(proj_input))
return self.average_pooling(proj_output)
消息传递神经网络(MPNN)
现在是完成 MPNN 模型的时候了。除了消息传递和读取,将实现一个两层分类网络来进行 BBBP 的预测。
def MPNNModel(
atom_dim,
bond_dim,
batch_size=32,
message_units=64,
message_steps=4,
num_attention_heads=8,
dense_units=512,
):
atom_features = layers.Input((atom_dim), dtype="float32", name="atom_features")
bond_features = layers.Input((bond_dim), dtype="float32", name="bond_features")
pair_indices = layers.Input((2), dtype="int32", name="pair_indices")
molecule_indicator = layers.Input((), dtype="int32", name="molecule_indicator")
x = MessagePassing(message_units, message_steps)(
[atom_features, bond_features, pair_indices]
)
x = TransformerEncoderReadout(
num_attention_heads, message_units, dense_units, batch_size
)([x, molecule_indicator])
x = layers.Dense(dense_units, activation="relu")(x)
x = layers.Dense(1, activation="sigmoid")(x)
model = keras.Model(
inputs=[atom_features, bond_features, pair_indices, molecule_indicator],
outputs=[x],
)
return model
mpnn = MPNNModel(
atom_dim=x_train[0][0][0].shape[0], bond_dim=x_train[1][0][0].shape[0],
)
mpnn.compile(
loss=keras.losses.BinaryCrossentropy(),
optimizer=keras.optimizers.Adam(learning_rate=5e-4),
metrics=[keras.metrics.AUC(name="AUC")],
)
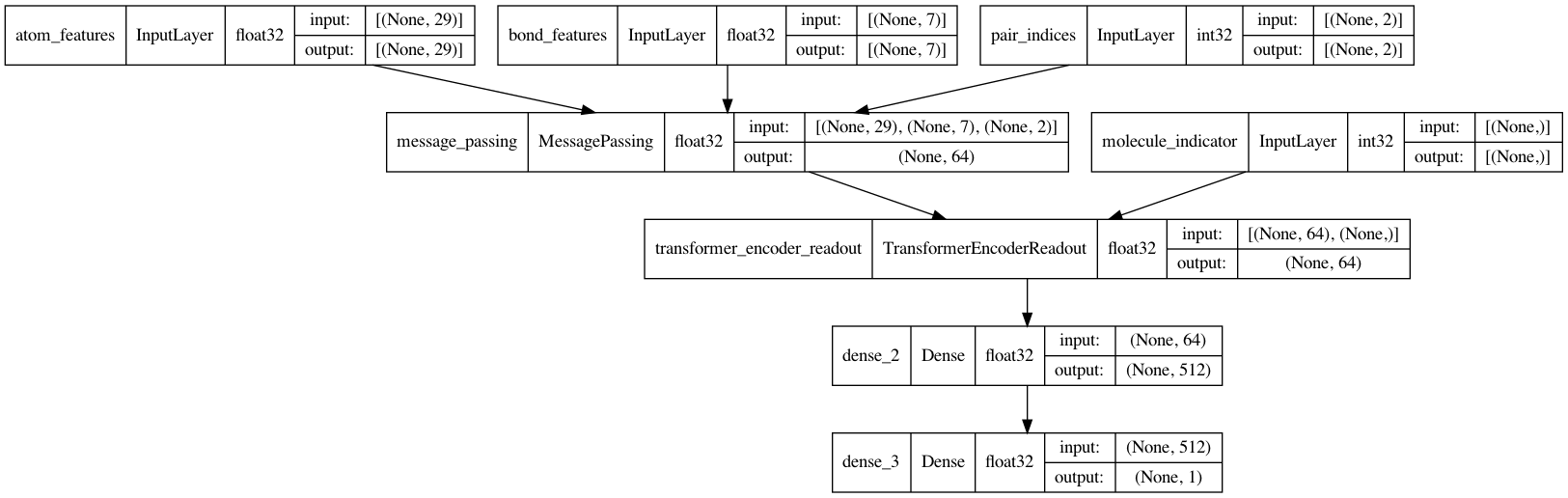
keras.utils.plot_model(mpnn, show_dtype=True, show_shapes=True)
训练
train_dataset = MPNNDataset(x_train, y_train)
valid_dataset = MPNNDataset(x_valid, y_valid)
test_dataset = MPNNDataset(x_test, y_test)
history = mpnn.fit(
train_dataset,
validation_data=valid_dataset,
epochs=40,
verbose=2,
class_weight={0: 2.0, 1: 0.5},
)
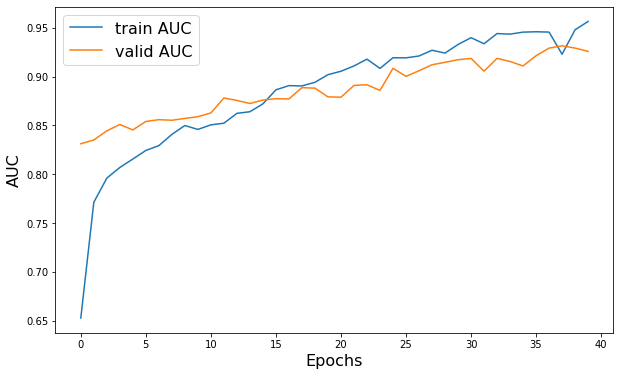
plt.figure(figsize=(10, 6))
plt.plot(history.history["AUC"], label="训练 AUC")
plt.plot(history.history["val_AUC"], label="验证 AUC")
plt.xlabel("轮次", fontsize=16)
plt.ylabel("AUC", fontsize=16)
plt.legend(fontsize=16)
Epoch 1/40 52/52 - 26s - loss: 0.5572 - AUC: 0.6527 - val_loss: 0.4660 - val_AUC: 0.8312 - 26s/epoch - 501ms/step Epoch 2/40 52/52 - 22s - loss: 0.4817 - AUC: 0.7713 - val_loss: 0.6889 - val_AUC: 0.8351 - 22s/epoch - 416ms/step Epoch 3/40 52/52 - 24s - loss: 0.4611 - AUC: 0.7960 - val_loss: 0.5863 - val_AUC: 0.8444 - 24s/epoch - 457ms/step Epoch 4/40 52/52 - 19s - loss: 0.4493 - AUC: 0.8069 - val_loss: 0.5059 - val_AUC: 0.8509 - 19s/epoch - 372ms/step Epoch 5/40 52/52 - 21s - loss: 0.4420 - AUC: 0.8155 - val_loss: 0.4965 - val_AUC: 0.8454 - 21s/epoch - 405ms/step Epoch 6/40 52/52 - 22s - loss: 0.4344 - AUC: 0.8243 - val_loss: 0.5307 - val_AUC: 0.8540 - 22s/epoch - 419ms/step Epoch 7/40 52/52 - 26s - loss: 0.4301 - AUC: 0.8293 - val_loss: 0.5131 - val_AUC: 0.8559 - 26s/epoch - 503ms/step Epoch 8/40 52/52 - 31s - loss: 0.4163 - AUC: 0.8408 - val_loss: 0.5361 - val_AUC: 0.8552 - 31s/epoch - 599ms/step Epoch 9/40 52/52 - 30s - loss: 0.4095 - AUC: 0.8499 - val_loss: 0.5371 - val_AUC: 0.8572 - 30s/epoch - 578ms/step Epoch 10/40 52/52 - 23s - loss: 0.4107 - AUC: 0.8459 - val_loss: 0.5923 - val_AUC: 0.8589 - 23s/epoch - 444ms/step Epoch 11/40 52/52 - 29s - loss: 0.4107 - AUC: 0.8505 - val_loss: 0.5070 - val_AUC: 0.8627 - 29s/epoch - 553ms/step Epoch 12/40 52/52 - 25s - loss: 0.4005 - AUC: 0.8522 - val_loss: 0.5417 - val_AUC: 0.8781 - 25s/epoch - 471ms/step Epoch 13/40 52/52 - 22s - loss: 0.3924 - AUC: 0.8623 - val_loss: 0.5915 - val_AUC: 0.8755 - 22s/epoch - 425ms/step Epoch 14/40 52/52 - 19s - loss: 0.3872 - AUC: 0.8640 - val_loss: 0.5852 - val_AUC: 0.8724 - 19s/epoch - 365ms/step Epoch 15/40 52/52 - 19s - loss: 0.3812 - AUC: 0.8720 - val_loss: 0.4949 - val_AUC: 0.8759 - 19s/epoch - 362ms/step Epoch 16/40 52/52 - 27s - loss: 0.3604 - AUC: 0.8864 - val_loss: 0.5076 - val_AUC: 0.8773 - 27s/epoch - 521ms/step Epoch 17/40 52/52 - 37s - loss: 0.3554 - AUC: 0.8907 - val_loss: 0.4556 - val_AUC: 0.8771 - 37s/epoch - 712ms/step Epoch 18/40 52/52 - 23s - loss: 0.3554 - AUC: 0.8904 - val_loss: 0.4854 - val_AUC: 0.8887 - 23s/epoch - 452ms/step Epoch 19/40 52/52 - 26s - loss: 0.3504 - AUC: 0.8942 - val_loss: 0.4622 - val_AUC: 0.8881 - 26s/epoch - 507ms/step Epoch 20/40 52/52 - 20s - loss: 0.3378 - AUC: 0.9019 - val_loss: 0.5568 - val_AUC: 0.8792 - 20s/epoch - 390ms/step Epoch 21/40 52/52 - 19s - loss: 0.3324 - AUC: 0.9055 - val_loss: 0.5623 - val_AUC: 0.8789 - 19s/epoch - 363ms/step Epoch 22/40 52/52 - 19s - loss: 0.3248 - AUC: 0.9109 - val_loss: 0.5486 - val_AUC: 0.8909 - 19s/epoch - 357ms/step Epoch 23/40 52/52 - 18s - loss: 0.3126 - AUC: 0.9179 - val_loss: 0.5684 - val_AUC: 0.8916 - 18s/epoch - 348ms/step Epoch 24/40 52/52 - 18s - loss: 0.3296 - AUC: 0.9084 - val_loss: 0.5462 - val_AUC: 0.8858 - 18s/epoch - 352ms/step Epoch 25/40 52/52 - 18s - loss: 0.3098 - AUC: 0.9193 - val_loss: 0.4212 - val_AUC: 0.9085 - 18s/epoch - 349ms/step Epoch 26/40 52/52 - 18s - loss: 0.3095 - AUC: 0.9192 - val_loss: 0.4991 - val_AUC: 0.9002 - 18s/epoch - 348ms/step Epoch 27/40 52/52 - 18s - loss: 0.3056 - AUC: 0.9211 - val_loss: 0.4739 - val_AUC: 0.9060 - 18s/epoch - 349ms/step Epoch 28/40 52/52 - 18s - loss: 0.2942 - AUC: 0.9270 - val_loss: 0.4188 - val_AUC: 0.9121 - 18s/epoch - 344ms/step Epoch 29/40 52/52 - 18s - loss: 0.3004 - AUC: 0.9241 - val_loss: 0.4056 - val_AUC: 0.9146 - 18s/epoch - 351ms/step Epoch 30/40 52/52 - 18s - loss: 0.2810 - AUC: 0.9328 - val_loss: 0.3923 - val_AUC: 0.9172 - 18s/epoch - 355ms/step Epoch 31/40 52/52 - 18s - loss: 0.2661 - AUC: 0.9398 - val_loss: 0.3609 - val_AUC: 0.9186 - 18s/epoch - 349ms/step Epoch 32/40 52/52 - 19s - loss: 0.2797 - AUC: 0.9336 - val_loss: 0.3764 - val_AUC: 0.9055 - 19s/epoch - 357ms/step Epoch 33/40 52/52 - 19s - loss: 0.2552 - AUC: 0.9441 - val_loss: 0.3941 - val_AUC: 0.9187 - 19s/epoch - 368ms/step Epoch 34/40 52/52 - 23s - loss: 0.2601 - AUC: 0.9435 - val_loss: 0.4128 - val_AUC: 0.9154 - 23s/epoch - 443ms/step Epoch 35/40 52/52 - 32s - loss: 0.2533 - AUC: 0.9455 - val_loss: 0.4191 - val_AUC: 0.9109 - 32s/epoch - 615ms/step Epoch 36/40 52/52 - 23s - loss: 0.2530 - AUC: 0.9459 - val_loss: 0.4276 - val_AUC: 0.9213 - 23s/epoch - 435ms/step Epoch 37/40 52/52 - 31s - loss: 0.2531 - AUC: 0.9456 - val_loss: 0.3950 - val_AUC: 0.9292 - 31s/epoch - 593ms/step Epoch 38/40 52/52 - 22s - loss: 0.3039 - AUC: 0.9229 - val_loss: 0.3114 - val_AUC: 0.9315 - 22s/epoch - 428ms/step Epoch 39/40 52/52 - 20s - loss: 0.2477 - AUC: 0.9479 - val_loss: 0.3584 - val_AUC: 0.9292 - 20s/epoch - 391ms/step Epoch 40/40 52/52 - 22s - loss: 0.2276 - AUC: 0.9565 - val_loss: 0.3279 - val_AUC: 0.9258 - 22s/epoch - 416ms/step
预测
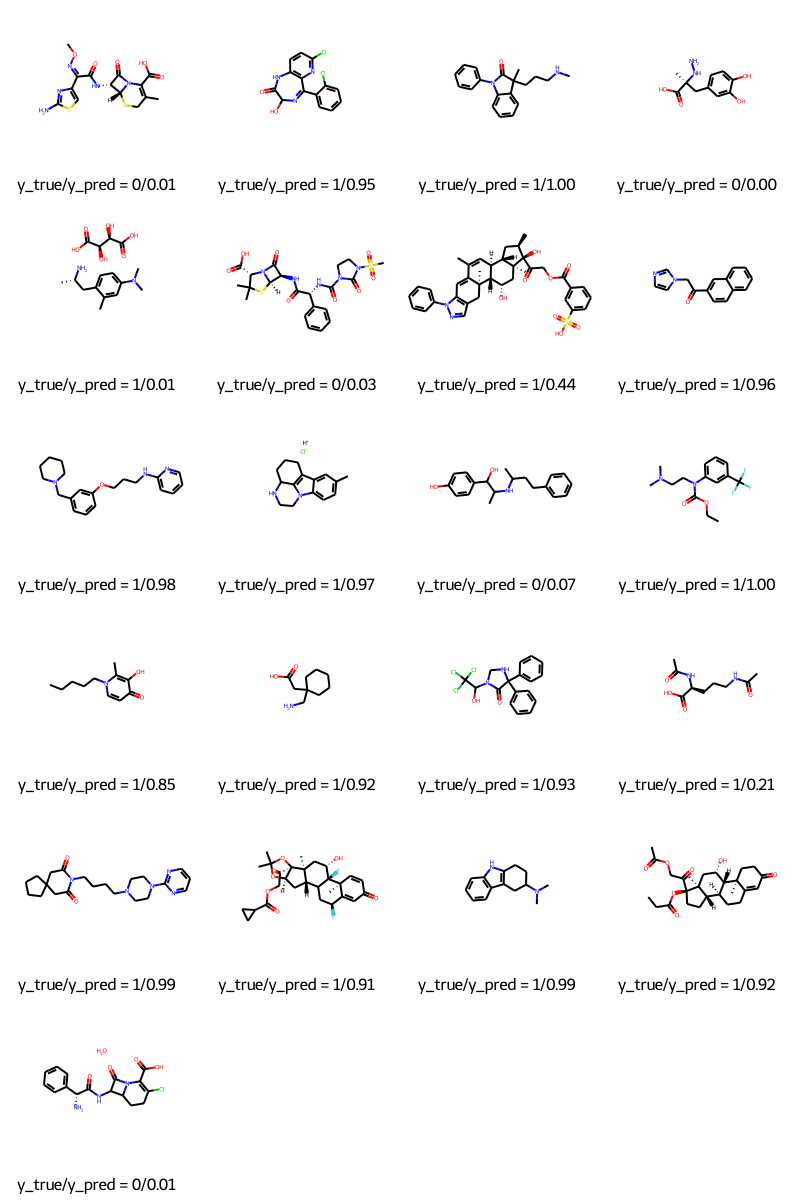
molecules = [molecule_from_smiles(df.smiles.values[index]) for index in test_index]
y_true = [df.p_np.values[index] for index in test_index]
y_pred = tf.squeeze(mpnn.predict(test_dataset), axis=1)
legends = [f"y_true/y_pred = {y_true[i]}/{y_pred[i]:.2f}" for i in range(len(y_true))]
MolsToGridImage(molecules, molsPerRow=4, legends=legends)
结论
在本教程中,我们演示了一种消息传递神经网络(MPNN)来预测多种不同分子的血脑屏障通透性(BBBP)。我们首先需要从SMILES构建图,然后构建一个可以操作这些图的Keras模型,最后训练模型以进行预测。
示例可在 HuggingFace 上找到
| 训练模型 | 演示 |
|---|---|
 |
 |
