! [ -e /content ] && pip install -Uqq fastai # 在Colab上升级fastai医学影像
用于处理DICOM文件的助手
::: {#cell-4 .cell 0=‘e’ 1=‘x’ 2=‘p’ 3=‘o’ 4=‘r’ 5=‘t’}
from __future__ import annotations
from fastai.basics import *
from fastai.vision.all import *
from fastai.data.transforms import *
import pydicom,kornia,skimage
from pydicom.dataset import Dataset as DcmDataset
from pydicom.tag import BaseTag as DcmTag
from pydicom.multival import MultiValue as DcmMultiValue
from PIL import Image
try:
import cv2
cv2.setNumThreads(0)
except: pass:::
from nbdev.showdoc import *matplotlib.rcParams['image.cmap'] = 'bone'_all_ = ['DcmDataset', 'DcmTag', 'DcmMultiValue', 'dcmread', 'get_dicom_files', 'DicomSegmentationDataLoaders']补丁修复
def get_dicom_files(path, recurse=True, folders=None):
"Get dicom files in `path` recursively, only in `folders`, if specified."
return get_files(path, extensions=[".dcm",".dicom"], recurse=recurse, folders=folders)@patch
def dcmread(fn:Path, force = False):
"Open a `DICOM` file"
return pydicom.dcmread(str(fn), force)fastai.medical.imaging 使用 pydicom.dcmread 来读取 DICOM 文件。要查看 DICOM 的 header,请指定测试文件的 path 并调用 dcmread。
TEST_DCM = Path('images/sample.dcm')
dcm = TEST_DCM.dcmread()
dcmDataset.file_meta -------------------------------
(0002, 0000) File Meta Information Group Length UL: 176
(0002, 0001) File Meta Information Version OB: b'\x00\x01'
(0002, 0002) Media Storage SOP Class UID UI: CT Image Storage
(0002, 0003) Media Storage SOP Instance UID UI: 9999.180975792154576730321054399332994563536
(0002, 0010) Transfer Syntax UID UI: Explicit VR Little Endian
(0002, 0012) Implementation Class UID UI: 1.2.40.0.13.1.1.1
(0002, 0013) Implementation Version Name SH: 'dcm4che-1.4.38'
-------------------------------------------------
(0008, 0018) SOP Instance UID UI: ID_e0cc6a4b5
(0008, 0060) Modality CS: 'CT'
(0010, 0020) Patient ID LO: 'ID_a107dd7f'
(0020, 000d) Study Instance UID UI: ID_6468bdd34a
(0020, 000e) Series Instance UID UI: ID_4be303ae64
(0020, 0010) Study ID SH: ''
(0020, 0032) Image Position (Patient) DS: [-125.000, -122.268, 115.936]
(0020, 0037) Image Orientation (Patient) DS: [1.000000, 0.000000, 0.000000, 0.000000, 0.978148, -0.207912]
(0028, 0002) Samples per Pixel US: 1
(0028, 0004) Photometric Interpretation CS: 'MONOCHROME2'
(0028, 0010) Rows US: 256
(0028, 0011) Columns US: 256
(0028, 0030) Pixel Spacing DS: [0.488281, 0.488281]
(0028, 0100) Bits Allocated US: 16
(0028, 0101) Bits Stored US: 16
(0028, 0102) High Bit US: 15
(0028, 0103) Pixel Representation US: 1
(0028, 1050) Window Center DS: "40.0"
(0028, 1051) Window Width DS: "100.0"
(0028, 1052) Rescale Intercept DS: "-1024.0"
(0028, 1053) Rescale Slope DS: "1.0"
(7fe0, 0010) Pixel Data OW: Array of 131072 elementstype(dcm)pydicom.dataset.FileDatasetclass TensorDicom(TensorImage):
"Inherits from `TensorImage` and converts the `pixel_array` into a `TensorDicom`"
_show_args = {'cmap':'gray'}class PILDicom(PILBase):
_open_args,_tensor_cls,_show_args = {},TensorDicom,TensorDicom._show_args
@classmethod
def create(cls, fn:Path|str|bytes, mode=None)->None:
"Open a `DICOM file` from path `fn` or bytes `fn` and load it as a `PIL Image`"
if isinstance(fn,bytes): im = Image.fromarray(pydicom.dcmread(pydicom.filebase.DicomBytesIO(fn)).pixel_array)
if isinstance(fn,(Path,str)): im = Image.fromarray(pydicom.dcmread(fn).pixel_array)
im.load()
im = im._new(im.im)
return cls(im.convert(mode) if mode else im)
PILDicom._tensor_cls = TensorDicom@patch
def png16read(self:Path): return array(Image.open(self), dtype=np.uint16)@patch(as_prop=True)
def pixels(self:DcmDataset):
"`pixel_array` as a tensor"
return tensor(self.pixel_array.astype(np.float32))dcm.pixelstensor([[-1024., -1024., -1024., ..., -1024., -1024., -1024.],
[-1024., -1024., -1024., ..., -1024., -1024., -1024.],
[-1024., -1024., -1024., ..., -1024., -1024., -1024.],
...,
[-1024., -1024., -1024., ..., -1024., -1024., -1024.],
[-1024., -1024., -1024., ..., -1024., -1024., -1024.],
[-1024., -1024., -1024., ..., -1024., -1024., -1024.]])@patch(as_prop=True)
def scaled_px(self:DcmDataset):
"`pixels` scaled by `RescaleSlope` and `RescaleIntercept`"
img = self.pixels
if hasattr(self, 'RescaleSlope') and hasattr(self, 'RescaleIntercept') is not None:
return img * self.RescaleSlope + self.RescaleIntercept
else: return imgscaled_px 使用 RescaleSlope 和 RescaleIntercept 值来正确缩放图像,使其能够表示正确的组织密度。您可以通过查看 dicom 图像的像素分布来观察 scaled_px 的作用。下方的直方图显示了当前的像素分布,显示像素范围在 -1133 和 2545 之间。
plt.hist(dcm.pixels.flatten().numpy());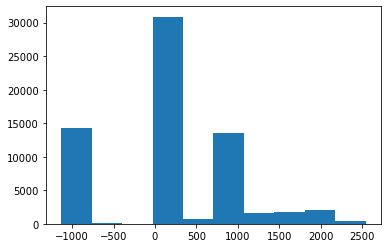
如测试图像的 header 所示,RescaleIntercept 的值为 -1024.0,而 RescaleSlope 的值为 1.0。scaled_px 将根据这些值对像素进行缩放。
plt.hist(dcm.scaled_px.flatten().numpy());
像素分布现在在 -2157 和 1521 之间。
def array_freqhist_bins(self, n_bins=100):
"A numpy based function to split the range of pixel values into groups, such that each group has around the same number of pixels"
imsd = np.sort(self.flatten())
t = np.array([0.001])
t = np.append(t, np.arange(n_bins)/n_bins+(1/2/n_bins))
t = np.append(t, 0.999)
t = (len(imsd)*t+0.5).astype(int)
return np.unique(imsd[t])@patch
def freqhist_bins(self:Tensor, n_bins=100):
"A function to split the range of pixel values into groups, such that each group has around the same number of pixels"
imsd = self.view(-1).sort()[0]
t = torch.cat([tensor([0.001]),
torch.arange(n_bins).float()/n_bins+(1/2/n_bins),
tensor([0.999])])
t = (len(imsd)*t).long()
return imsd[t].unique()例如,设置 n_bins 为 1 意味着这些区间将分成 3 个不同的区间(开始、结束和 n_bins 指定的区间数)。
t_bin = dcm.pixels.freqhist_bins(n_bins=1)
t_bintensor([-1076., 40., 2375.])plt.hist(t_bin.numpy(), bins=t_bin, color='c')
plt.plot(t_bin, torch.linspace(0,1,len(t_bin)));
使用 n_bins 设置为 100
t_bin = dcm.pixels.freqhist_bins(n_bins=100)
t_bintensor([-1076., -1026., -1024., -1021., 28., 30., 31., 32., 33.,
34., 35., 36., 37., 38., 39., 40., 41., 42.,
44., 48., 52., 58., 66., 72., 76., 80., 85.,
91., 94., 98., 103., 111., 123., 161., 219., 478.,
829., 999., 1027., 1038., 1044., 1047., 1049., 1050., 1051.,
1052., 1053., 1054., 1055., 1056., 1057., 1058., 1059., 1060.,
1062., 1066., 1108., 1265., 1453., 1616., 1741., 1838., 1943.,
2051., 2220., 2375.])plt.hist(t_bin.numpy(), bins=t_bin, color='c'); plt.plot(t_bin, torch.linspace(0,1,len(t_bin)));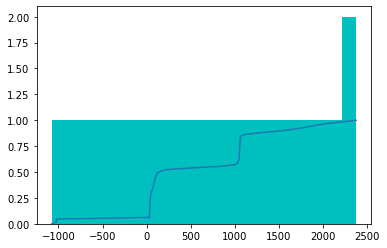
@patch
def hist_scaled_pt(self:Tensor, brks=None):
# 仅限 Pytorch 版本 - 当 interp_1d 能够优化时,切换到此版本。
if brks is None: brks = self.freqhist_bins()
brks = brks.to(self.device)
ys = torch.linspace(0., 1., len(brks)).to(self.device)
return self.flatten().interp_1d(brks, ys).reshape(self.shape).clamp(0.,1.)@patch
def hist_scaled(self:Tensor, brks=None):
"Scales a tensor using `freqhist_bins` to values between 0 and 1"
if self.device.type=='cuda': return self.hist_scaled_pt(brks)
if brks is None: brks = self.freqhist_bins()
ys = np.linspace(0., 1., len(brks))
x = self.numpy().flatten()
x = np.interp(x, brks.numpy(), ys)
return tensor(x).reshape(self.shape).clamp(0.,1.)测试图像的像素值范围在 -1000 和 2500 之间。
plt.hist(dcm.pixels.flatten().numpy(), bins=100);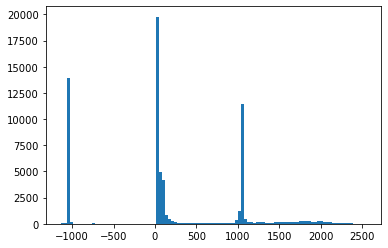
hist_scaled 提供了一种将输入像素值缩放到 0 和 1 之间的方法。
tensor_hists = dcm.pixels.hist_scaled()
plt.hist(tensor_hists.flatten().numpy(), bins=100);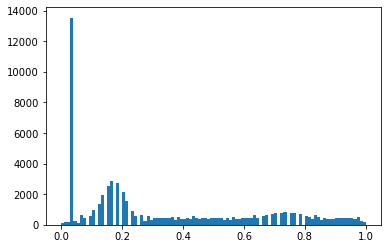
@patch
def hist_scaled(self:DcmDataset, brks=None, min_px=None, max_px=None):
"Pixels scaled to a `min_px` and `max_px` value"
px = self.scaled_px
if min_px is not None: px[px<min_px] = min_px
if max_px is not None: px[px>max_px] = max_px
return px.hist_scaled(brks=brks)data_scaled = dcm.hist_scaled()
plt.imshow(data_scaled, cmap=plt.cm.bone);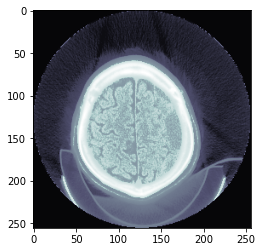
data_scaled = dcm.hist_scaled(min_px=100, max_px=1000)
plt.imshow(data_scaled, cmap=plt.cm.bone);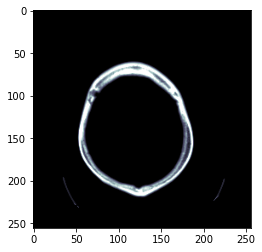
Dicom图像可以包含大量的体素值,而窗宽调整可以被视为一种操控这些值的手段,以便改变图像的外观,从而突出特定的结构。一个窗有两个值:
l= 窗位或中心,也称为亮度w= 窗宽或范围,也称为对比度
@patch
def windowed(self:Tensor, w, l):
"Scale pixel intensity by window width and window level"
px = self.clone()
px_min = l - w//2
px_max = l + w//2
px[px<px_min] = px_min
px[px>px_max] = px_max
return (px-px_min) / (px_max-px_min)@patch
def windowed(self:DcmDataset, w, l):
return self.scaled_px.windowed(w,l)# 来自 https://radiopaedia.org/articles/windowing-ct 的段落:
窗口化(CT)
在计算机断层扫描(CT)成像中,窗口化是一种用于调整图像显示的技术,以便更好地观察特定组织或病理特征。它涉及选择适当的窗口宽度(WW)和窗口中心(WL),以优化图像对比度和亮度,从而使感兴趣的结构在视觉上更加突出。
窗口宽度决定了图像中显示的灰度范围,而窗口中心则决定了该范围的中心点。通过调整这两个参数,放射科医生和其他医学专业人员可以更有效地识别和分析不同类型的组织和病变。
例如,软组织窗口通常用于观察肌肉、脂肪和内脏器官,而骨窗口则用于清晰显示骨骼结构。肺部窗口则专门用于评估肺实质和气道。
窗口化是CT图像解读中的一个重要工具,能够显著提高诊断的准确性和效率。
dicom_windows = types.SimpleNamespace(
brain=(80,40),
subdural=(254,100),
stroke=(8,32),
brain_bone=(2800,600),
brain_soft=(375,40),
lungs=(1500,-600),
mediastinum=(350,50),
abdomen_soft=(400,50),
liver=(150,30),
spine_soft=(250,50),
spine_bone=(1800,400)
)plt.imshow(dcm.windowed(*dicom_windows.brain), cmap=plt.cm.bone);
class TensorCTScan(TensorImageBW):
"Inherits from `TensorImageBW` and converts the `pixel_array` into a `TensorCTScan`"
_show_args = {'cmap':'bone'}tensor_ct = TensorCTScan(dcm.pixel_array)
tensor_ct.show();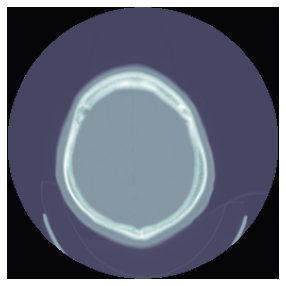
class PILCTScan(PILBase): _open_args,_tensor_cls,_show_args = {},TensorCTScan,TensorCTScan._show_args@patch
@delegates(show_image)
def show(self:DcmDataset, scale=True, cmap=plt.cm.bone, min_px=-1100, max_px=None, **kwargs):
"Display a normalized dicom image by default"
px = (self.windowed(*scale) if isinstance(scale,tuple)
else self.hist_scaled(min_px=min_px,max_px=max_px,brks=scale) if isinstance(scale,(ndarray,Tensor))
else self.hist_scaled(min_px=min_px,max_px=max_px) if scale
else self.scaled_px)
show_image(px, cmap=cmap, **kwargs)scales = False, True, dicom_windows.brain, dicom_windows.subdural
titles = 'raw','normalized','brain windowed','subdural windowed'
for s,a,t in zip(scales, subplots(2,2,imsize=4)[1].flat, titles):
dcm.show(scale=s, ax=a, title=t)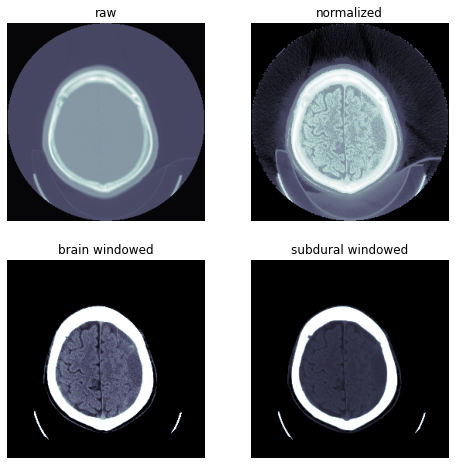
dcm.show(cmap=plt.cm.gist_ncar, figsize=(6,6))
一些dicom数据集,例如甲状腺超声分割数据集,是一个每个文件包含多个帧(在这种情况下是数百帧)的数据集。默认情况下,show函数将显示1帧,但如果数据集包含多个帧,您可以指定要查看的帧数。
@patch
def show(self:DcmDataset, frames=1, scale=True, cmap=plt.cm.bone, min_px=-1100, max_px=None, **kwargs):
"Adds functionality to view dicom images where each file may have more than 1 frame"
px = (self.windowed(*scale) if isinstance(scale,tuple)
else self.hist_scaled(min_px=min_px,max_px=max_px,brks=scale) if isinstance(scale,(ndarray,Tensor))
else self.hist_scaled(min_px=min_px,max_px=max_px) if scale
else self.scaled_px)
if px.ndim > 2:
gh=[]
p = px.shape; print(f'{p[0]} frames per file')
for i in range(frames): u = px[i]; gh.append(u)
show_images(gh, **kwargs)
else: show_image(px, cmap=cmap, **kwargs)dcm.show()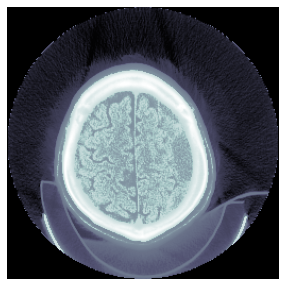
@patch
def pct_in_window(dcm:DcmDataset, w, l):
"% of pixels in the window `(w,l)`"
px = dcm.scaled_px
return ((px > l-w//2) & (px < l+w//2)).float().mean().item()dcm.pct_in_window(*dicom_windows.brain)0.19049072265625pct_in_window 可以用来检查图像中有多少百分比的像素是由有意义的像素组成的(在指定窗口内的像素)
def uniform_blur2d(x,s):
"Uniformly apply blurring"
w = x.new_ones(1,1,1,s)/s
# 将二维卷积分解为两个一维卷积
x = unsqueeze(x, dim=0, n=4-x.dim())
r = (F.conv2d(x, w, padding=s//2))
r = (F.conv2d(r, w.transpose(-1,-2), padding=s//2)).cpu()[:,0]
return r.squeeze()ims = dcm.hist_scaled(), uniform_blur2d(dcm.hist_scaled(), 20), uniform_blur2d(dcm.hist_scaled(), 50)
show_images(ims, titles=('original', 'blurred 20', 'blurred 50'))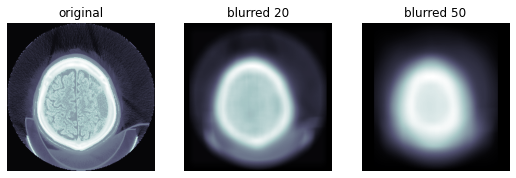
def gauss_blur2d(x,s):
"Apply gaussian_blur2d kornia filter"
s2 = int(s/4)*2+1
x2 = unsqueeze(x, dim=0, n=4-x.dim())
res = kornia.filters.gaussian_blur2d(x2, (s2,s2), (s,s), 'replicate')
return res.squeeze()ims = dcm.hist_scaled(), gauss_blur2d(dcm.hist_scaled(), 20), gauss_blur2d(dcm.hist_scaled(), 50)
show_images(ims, titles=('original', 'gauss_blur 20', 'gauss_blur 50'))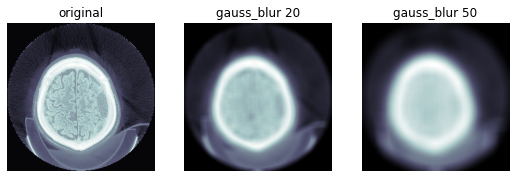
图像通常受到强度值随机变化的影响,这被称为噪声。高斯噪声包含从高斯或正态分布中抽取的强度变化。高斯滤波器通常用于模糊边缘并去除较小或较薄的区域,以保留最重要的信息。
@patch
def mask_from_blur(x:Tensor, window, sigma=0.3, thresh=0.05, remove_max=True):
"Create a mask from the blurred image"
p = x.windowed(*window)
if remove_max: p[p==1] = 0
return gauss_blur2d(p, s=sigma*x.shape[-1])>thresh@patch
def mask_from_blur(x:DcmDataset, window, sigma=0.3, thresh=0.05, remove_max=True):
"Create a mask from the blurred image"
return to_device(x.scaled_px).mask_from_blur(window, sigma, thresh, remove_max=remove_max)mask = dcm.mask_from_blur(dicom_windows.brain, sigma=0.9, thresh=0.1, remove_max=True)
wind = dcm.windowed(*dicom_windows.brain)
_,ax = subplots(1,3)
show_image(wind, ax=ax[0], title='window')
show_image(mask, alpha=0.5, cmap=plt.cm.Reds, ax=ax[1], title='mask')
show_image(wind, ax=ax[2])
show_image(mask, alpha=0.5, cmap=plt.cm.Reds, ax=ax[2], title='window and mask');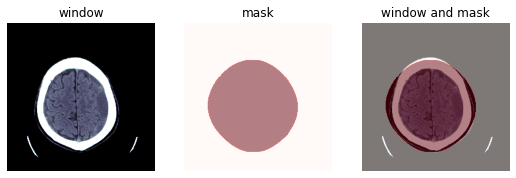
def _px_bounds(x, dim):
c = x.sum(dim).nonzero().cpu()
idxs,vals = torch.unique(c[:,0],return_counts=True)
vs = torch.split_with_sizes(c[:,1],tuple(vals))
d = {k.item():v for k,v in zip(idxs,vs)}
default_u = tensor([0,x.shape[-1]-1])
b = [d.get(o,default_u) for o in range(x.shape[0])]
b = [tensor([o.min(),o.max()]) for o in b]
return torch.stack(b)def mask2bbox(mask):
no_batch = mask.dim()==2
if no_batch: mask = mask[None]
bb1 = _px_bounds(mask,-1).t()
bb2 = _px_bounds(mask,-2).t()
res = torch.stack([bb1,bb2],dim=1).to(mask.device)
return res[...,0] if no_batch else resbbs = mask2bbox(mask)
lo,hi = bbs
show_image(wind[lo[0]:hi[0],lo[1]:hi[1]]);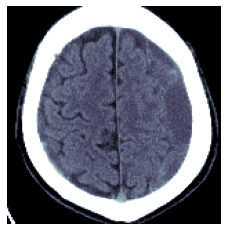
def _bbs2sizes(crops, init_sz, use_square=True):
bb = crops.flip(1)
szs = (bb[1]-bb[0])
if use_square: szs = szs.max(0)[0][None].repeat((2,1))
overs = (szs+bb[0])>init_sz
bb[0][overs] = init_sz-szs[overs]
lows = (bb[0]/float(init_sz))
return lows,szs/float(init_sz)def crop_resize(x, crops, new_sz):
# NB 假设输入为方形。未对任何非方形输入进行测试!
bs = x.shape[0]
lows,szs = _bbs2sizes(crops, x.shape[-1])
if not isinstance(new_sz,(list,tuple)): new_sz = (new_sz,new_sz)
id_mat = tensor([[1.,0,0],[0,1,0]])[None].repeat((bs,1,1)).to(x.device)
with warnings.catch_warnings():
warnings.filterwarnings('ignore', category=UserWarning)
sp = F.affine_grid(id_mat, (bs,1,*new_sz))+1.
grid = sp*unsqueeze(szs.t(),1,n=2)+unsqueeze(lows.t()*2.,1,n=2)
return F.grid_sample(x.unsqueeze(1), grid-1)px256 = crop_resize(to_device(wind[None]), bbs[...,None], 128)[0]
show_image(px256)
px256.shapetorch.Size([1, 128, 128])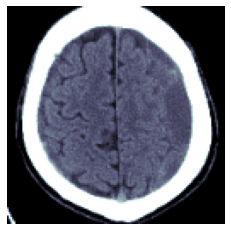
比较原始图像与使用mask和crop_resize函数处理后的图像
_,axs = subplots(1,2)
dcm.show(ax=axs[0])
show_image(px256, ax=axs[1]);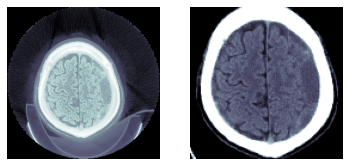
@patch
def to_nchan(x:Tensor, wins, bins=None):
res = [x.windowed(*win) for win in wins]
if not isinstance(bins,int) or bins!=0: res.append(x.hist_scaled(bins).clamp(0,1))
dim = [0,1][x.dim()==3]
return TensorCTScan(torch.stack(res, dim=dim))@patch
def to_nchan(x:DcmDataset, wins, bins=None):
return x.scaled_px.to_nchan(wins, bins)to_nchan 接收一个张量或 dicom 作为输入,并返回多个单通道图像(第一个取决于选择的 windows 和一个归一化的图像)。将 bins 设置为 0 仅返回窗口化图像。
show_images(dcm.to_nchan([dicom_windows.brain], bins=0))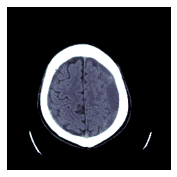
show_images(dcm.to_nchan([dicom_windows.brain], bins=None))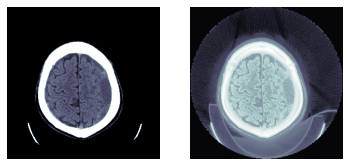
@patch
def to_3chan(x:Tensor, win1, win2, bins=None):
return x.to_nchan([win1,win2],bins=bins)@patch
def to_3chan(x:DcmDataset, win1, win2, bins=None):
return x.scaled_px.to_3chan(win1, win2, bins)show_images(dcm.to_nchan([dicom_windows.brain,dicom_windows.subdural,dicom_windows.abdomen_soft]))
@patch
def save_jpg(x:Tensor|DcmDataset, path, wins, bins=None, quality=90):
"Save tensor or dicom image into `jpg` format"
fn = Path(path).with_suffix('.jpg')
x = (x.to_nchan(wins, bins)*255).byte()
im = Image.fromarray(x.permute(1,2,0).numpy(), mode=['RGB','CMYK'][x.shape[0]==4])
im.save(fn, quality=quality)@patch
def to_uint16(x:Tensor|DcmDataset, bins=None):
"Convert into a unit16 array"
d = x.hist_scaled(bins).clamp(0,1) * 2**16
return d.numpy().astype(np.uint16)@patch
def save_tif16(x:Tensor|DcmDataset, path, bins=None, compress=True):
"Save tensor or dicom image into `tiff` format"
fn = Path(path).with_suffix('.tif')
Image.fromarray(x.to_uint16(bins)).save(str(fn), compression='tiff_deflate' if compress else None)_,axs=subplots(1,2)
with tempfile.TemporaryDirectory() as f:
f = Path(f)
dcm.save_jpg(f/'test.jpg', [dicom_windows.brain,dicom_windows.subdural])
show_image(Image.open(f/'test.jpg'), ax=axs[0])
dcm.save_tif16(f/'test.tif')
show_image(Image.open(str(f/'test.tif')), ax=axs[1]);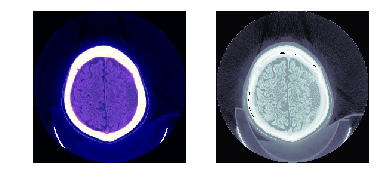
@patch
def set_pixels(self:DcmDataset, px):
self.PixelData = px.tobytes()
self.Rows,self.Columns = px.shape
DcmDataset.pixel_array = property(DcmDataset.pixel_array.fget, set_pixels)@patch
def zoom(self:DcmDataset, ratio):
"Zoom image by specified ratio"
with warnings.catch_warnings():
warnings.simplefilter("ignore", UserWarning)
self.set_pixels(ndimage.zoom(self.pixel_array, ratio))检查DICOM图像的当前大小
dcm.pixel_array.shape(256, 256)dcm.zoom(7.0)
dcm.show(); dcm.pixel_array.shape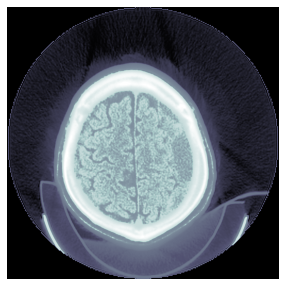
@patch
def zoom_to(self:DcmDataset, sz):
"Change image size to specified pixel size"
if not isinstance(sz,(list,tuple)): sz=(sz,sz)
rows,cols = sz
self.zoom((rows/self.Rows,cols/self.Columns))dcm.zoom_to(200); dcm.pixel_array.shape(200, 200)@patch(as_prop=True)
def shape(self:DcmDataset):
"Returns the shape of a dicom image as rows and columns"
return self.Rows,self.Columnsdcm2 = TEST_DCM.dcmread()
dcm2.zoom_to(90)
test_eq(dcm2.shape, (90,90))dcm2 = TEST_DCM.dcmread()
dcm2.zoom(0.25)
dcm2.show()
def _cast_dicom_special(x):
cls = type(x)
if not cls.__module__.startswith('pydicom'): return x
if cls.__base__ == object: return x
return cls.__base__(x)
def _split_elem(vals):
res = dict()
for val in vals:
k, v = val.keyword, val.value
if not isinstance(v,DcmMultiValue):
res[k] = v
continue
res[f'Multi{k}'] = 1
for i,o in enumerate(v): res[f'{k}{"" if i==0 else i}'] = o
return {k: _cast_dicom_special(v) for k, v in res.items()}@patch
def as_dict(self:DcmDataset, px_summ=True, window=dicom_windows.brain):
"Convert the header of a dicom into a dictionary"
pxdata = (0x7fe0,0x0010)
vals = [self[o] for o in self.keys() if o != pxdata]
res = _split_elem(vals)
res['fname'] = self.filename
if not px_summ: return res
stats = 'min','max','mean','std'
try:
pxs = self.pixel_array
for f in stats: res['img_'+f] = getattr(pxs,f)()
res['img_pct_window'] = self.pct_in_window(*window)
except Exception as e:
for f in stats: res['img_'+f] = 0
print(res,e)
return resas_dict 接受两个参数:px_summ,默认设置为 True,它将返回额外的统计信息,例如最小像素值、最大像素值、平均像素值和图像标准差。 window 参数根据指定的 window 计算 pct_in_window 的值。
dcm.as_dict(px_summ=True, window=dicom_windows.brain);def _dcm2dict(fn, window=dicom_windows.brain, px_summ=True, **kwargs):
return fn.dcmread().as_dict(window=window, px_summ=px_summ, **kwargs)@delegates(parallel)
def _from_dicoms(cls, fns, n_workers=0, **kwargs):
return pd.DataFrame(parallel(_dcm2dict, fns, n_workers=n_workers, **kwargs))
pd.DataFrame.from_dicoms = classmethod(_from_dicoms)创建一个包含dicom的header值的数据框
pneumothorax_source = untar_data(URLs.SIIM_SMALL)
items = get_dicom_files(pneumothorax_source, recurse=True, folders='train')
dicom_dataframe = pd.DataFrame.from_dicoms(items, window=dicom_windows.brain, px_summ=True)
dicom_dataframe.head(2).T.tail(5)| 0 | 1 | |
|---|---|---|
| img_min | 0 | 0 |
| img_max | 254 | 250 |
| img_mean | 160.398 | 114.525 |
| img_std | 53.8549 | 70.7523 |
| img_pct_window | 0.0870295 | 0.326269 |
class DicomSegmentationDataLoaders(DataLoaders):
"Basic wrapper around DICOM `DataLoaders` with factory methods for segmentation problems"
@classmethod
@delegates(DataLoaders.from_dblock)
def from_label_func(cls, path, fnames, label_func, valid_pct=0.2, seed=None, codes=None, item_tfms=None, batch_tfms=None, **kwargs):
"Create from list of `fnames` in `path`s with `label_func`."
dblock = DataBlock(blocks=(ImageBlock(cls=PILDicom), MaskBlock(codes=codes)),
splitter=RandomSplitter(valid_pct, seed=seed),
get_y=label_func,
item_tfms=item_tfms,
batch_tfms=batch_tfms)
res = cls.from_dblock(dblock, fnames, path=path, **kwargs)
return respath = untar_data(URLs.TCGA_SMALL)
codes = np.loadtxt(path/'codes.txt', dtype=str)
fnames = get_dicom_files(path/'dicoms')
label_func = lambda o: path/'labels'/f'{o.stem}.png'
dls = DicomSegmentationDataLoaders.from_label_func(path, fnames, label_func, codes=codes, bs=4)
dls.show_batch()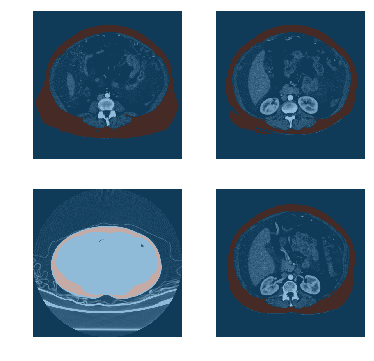
导出 -
from nbdev import nbdev_export
nbdev_export()Converted 00_torch_core.ipynb.
Converted 01_layers.ipynb.
Converted 01a_losses.ipynb.
Converted 02_data.load.ipynb.
Converted 03_data.core.ipynb.
Converted 04_data.external.ipynb.
Converted 05_data.transforms.ipynb.
Converted 06_data.block.ipynb.
Converted 07_vision.core.ipynb.
Converted 08_vision.data.ipynb.
Converted 09_vision.augment.ipynb.
Converted 09b_vision.utils.ipynb.
Converted 09c_vision.widgets.ipynb.
Converted 10_tutorial.pets.ipynb.
Converted 10b_tutorial.albumentations.ipynb.
Converted 11_vision.models.xresnet.ipynb.
Converted 12_optimizer.ipynb.
Converted 13_callback.core.ipynb.
Converted 13a_learner.ipynb.
Converted 13b_metrics.ipynb.
Converted 14_callback.schedule.ipynb.
Converted 14a_callback.data.ipynb.
Converted 15_callback.hook.ipynb.
Converted 15a_vision.models.unet.ipynb.
Converted 16_callback.progress.ipynb.
Converted 17_callback.tracker.ipynb.
Converted 18_callback.fp16.ipynb.
Converted 18a_callback.training.ipynb.
Converted 18b_callback.preds.ipynb.
Converted 19_callback.mixup.ipynb.
Converted 20_interpret.ipynb.
Converted 20a_distributed.ipynb.
Converted 21_vision.learner.ipynb.
Converted 22_tutorial.imagenette.ipynb.
Converted 23_tutorial.vision.ipynb.
Converted 24_tutorial.siamese.ipynb.
Converted 24_vision.gan.ipynb.
Converted 30_text.core.ipynb.
Converted 31_text.data.ipynb.
Converted 32_text.models.awdlstm.ipynb.
Converted 33_text.models.core.ipynb.
Converted 34_callback.rnn.ipynb.
Converted 35_tutorial.wikitext.ipynb.
Converted 36_text.models.qrnn.ipynb.
Converted 37_text.learner.ipynb.
Converted 38_tutorial.text.ipynb.
Converted 39_tutorial.transformers.ipynb.
Converted 40_tabular.core.ipynb.
Converted 41_tabular.data.ipynb.
Converted 42_tabular.model.ipynb.
Converted 43_tabular.learner.ipynb.
Converted 44_tutorial.tabular.ipynb.
Converted 45_collab.ipynb.
Converted 46_tutorial.collab.ipynb.
Converted 50_tutorial.datablock.ipynb.
Converted 60_medical.imaging.ipynb.
Converted 61_tutorial.medical_imaging.ipynb.
Converted 65_medical.text.ipynb.
Converted 70_callback.wandb.ipynb.
Converted 71_callback.tensorboard.ipynb.
Converted 72_callback.neptune.ipynb.
Converted 73_callback.captum.ipynb.
Converted 74_callback.azureml.ipynb.
Converted 97_test_utils.ipynb.
Converted 99_pytorch_doc.ipynb.
Converted dev-setup.ipynb.
Converted index.ipynb.
Converted quick_start.ipynb.
Converted tutorial.ipynb.