Note
Go to the end to download the full example code. or to run this example in your browser via Binder
变分贝叶斯高斯混合模型的浓度先验类型分析#
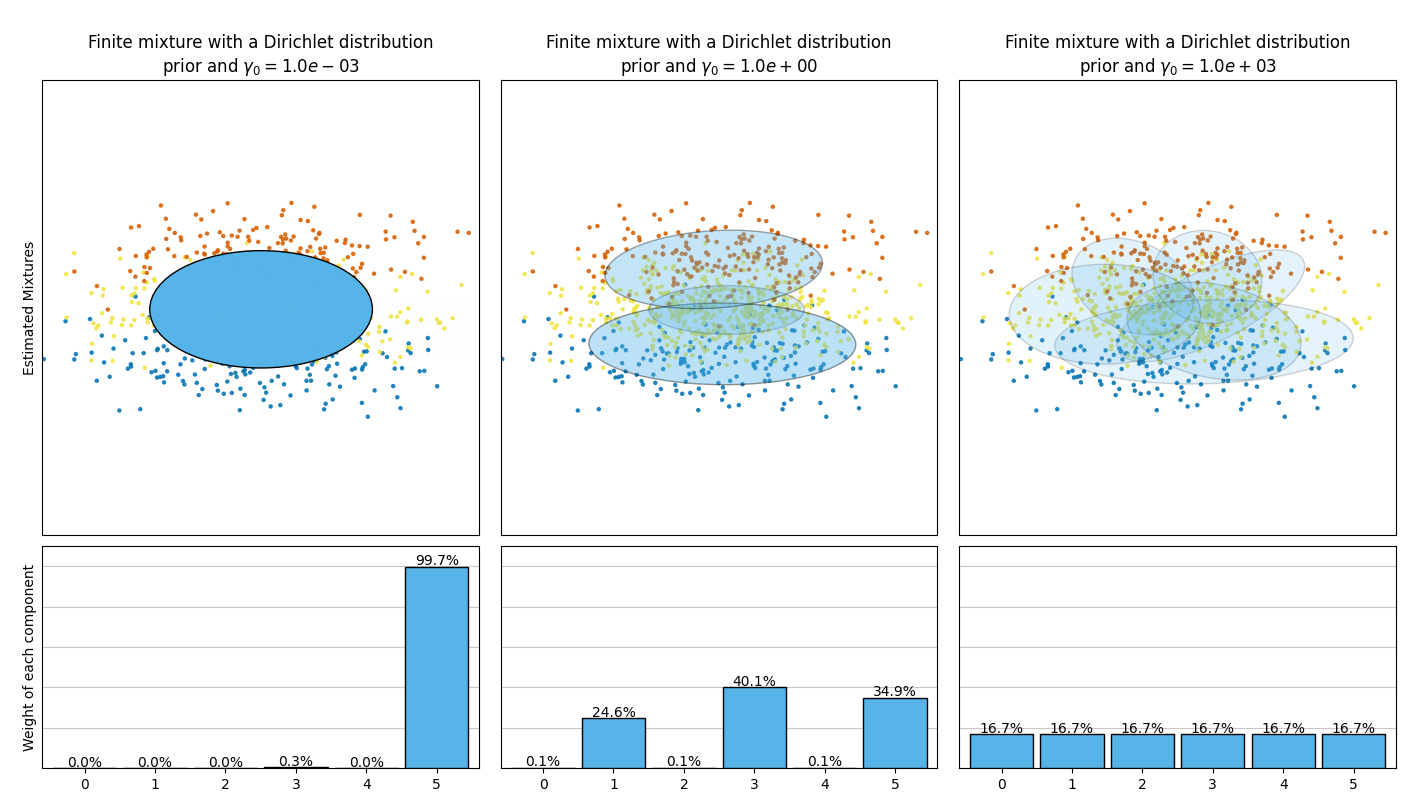
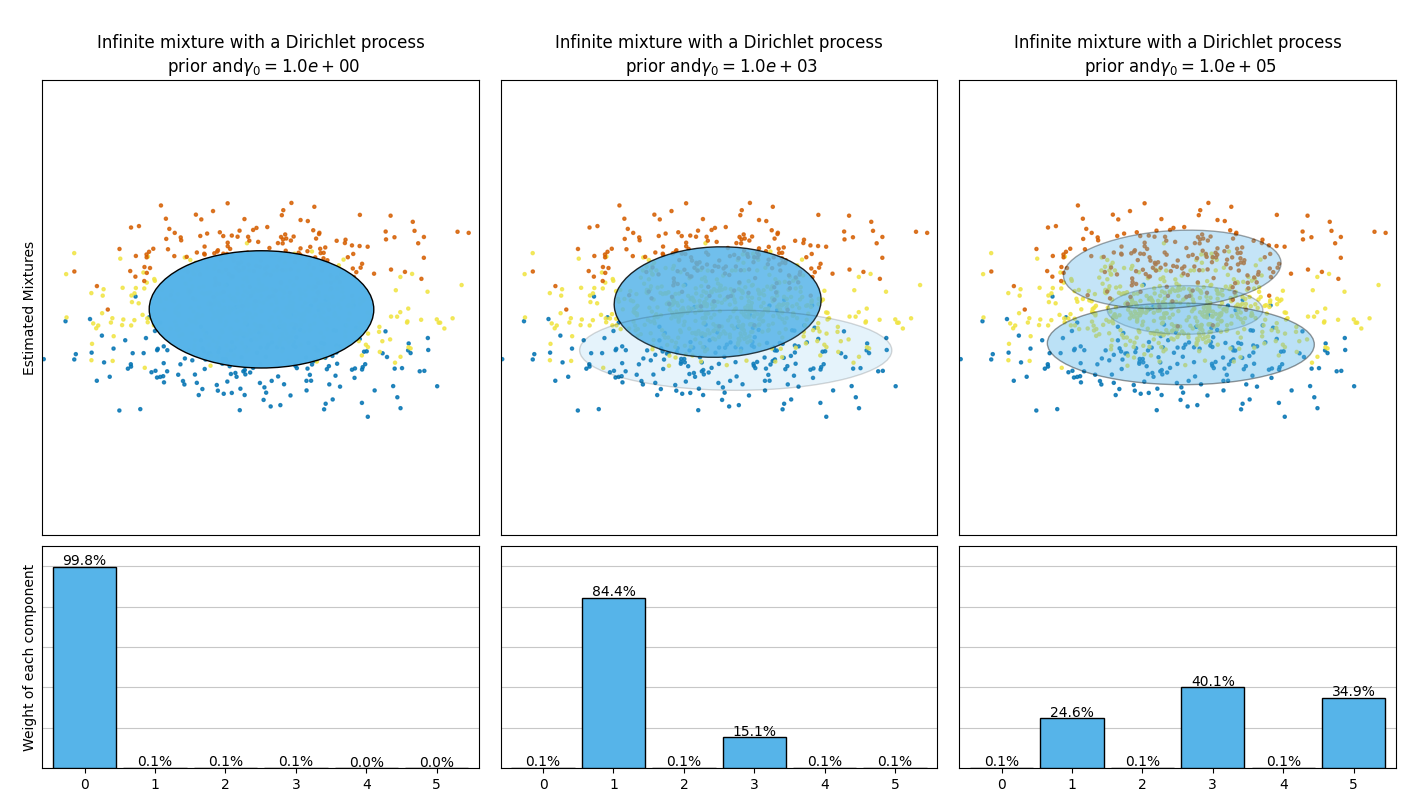
本示例绘制了通过 BayesianGaussianMixture 类模型拟合的玩具数据集(三个高斯分布的混合)得到的椭圆体图。模型使用了狄利克雷分布先验( weight_concentration_prior_type='dirichlet_distribution' )和狄利克雷过程先验( weight_concentration_prior_type='dirichlet_process' )。在每个图中,我们绘制了三种不同权重浓度先验值的结果。
BayesianGaussianMixture类可以自动调整其混合成分的数量。参数weight_concentration_prior与具有非零权重的成分数量直接相关。为浓度先验指定较低的值会使模型将大部分权重放在少数成分上,并将其余成分的权重设定得非常接近于零。较高的浓度先验值将允许更多的成分在混合中处于活跃状态。
狄利克雷过程先验允许定义无限数量的成分,并自动选择正确的成分数量:仅在必要时激活一个成分。
相反,具有狄利克雷分布先验的经典有限混合模型将更倾向于均匀加权的成分,因此往往会将自然聚类划分为不必要的子成分。
# 作者:scikit-learn 开发者
# SPDX 许可证标识符:BSD-3-Clause
import matplotlib as mpl
import matplotlib.gridspec as gridspec
import matplotlib.pyplot as plt
import numpy as np
from sklearn.mixture import BayesianGaussianMixture
def plot_ellipses(ax, weights, means, covars):
for n in range(means.shape[0]):
eig_vals, eig_vecs = np.linalg.eigh(covars[n])
unit_eig_vec = eig_vecs[0] / np.linalg.norm(eig_vecs[0])
angle = np.arctan2(unit_eig_vec[1], unit_eig_vec[0])
# 椭圆需要度数
angle = 180 * angle / np.pi
# 特征向量归一化
eig_vals = 2 * np.sqrt(2) * np.sqrt(eig_vals)
ell = mpl.patches.Ellipse(
means[n], eig_vals[0], eig_vals[1], angle=180 + angle, edgecolor="black"
)
ell.set_clip_box(ax.bbox)
ell.set_alpha(weights[n])
ell.set_facecolor("#56B4E9")
ax.add_artist(ell)
def plot_results(ax1, ax2, estimator, X, y, title, plot_title=False):
ax1.set_title(title)
ax1.scatter(X[:, 0], X[:, 1], s=5, marker="o", color=colors[y], alpha=0.8)
ax1.set_xlim(-2.0, 2.0)
ax1.set_ylim(-3.0, 3.0)
ax1.set_xticks(())
ax1.set_yticks(())
plot_ellipses(ax1, estimator.weights_, estimator.means_, estimator.covariances_)
ax2.get_xaxis().set_tick_params(direction="out")
ax2.yaxis.grid(True, alpha=0.7)
for k, w in enumerate(estimator.weights_):
ax2.bar(
k,
w,
width=0.9,
color="#56B4E9",
zorder=3,
align="center",
edgecolor="black",
)
ax2.text(k, w + 0.007, "%.1f%%" % (w * 100.0), horizontalalignment="center")
ax2.set_xlim(-0.6, 2 * n_components - 0.4)
ax2.set_ylim(0.0, 1.1)
ax2.tick_params(axis="y", which="both", left=False, right=False, labelleft=False)
ax2.tick_params(axis="x", which="both", top=False)
if plot_title:
ax1.set_ylabel("Estimated Mixtures")
ax2.set_ylabel("Weight of each component")
# 数据集的参数
random_state, n_components, n_features = 2, 3, 2
colors = np.array(["#0072B2", "#F0E442", "#D55E00"])
covars = np.array(
[[[0.7, 0.0], [0.0, 0.1]], [[0.5, 0.0], [0.0, 0.1]], [[0.5, 0.0], [0.0, 0.1]]]
)
samples = np.array([200, 500, 200])
means = np.array([[0.0, -0.70], [0.0, 0.0], [0.0, 0.70]])
# mean_precision_prior= 0.8 以最小化先验的影响
estimators = [
(
"Finite mixture with a Dirichlet distribution\nprior and " r"$\gamma_0=$",
BayesianGaussianMixture(
weight_concentration_prior_type="dirichlet_distribution",
n_components=2 * n_components,
reg_covar=0,
init_params="random",
max_iter=1500,
mean_precision_prior=0.8,
random_state=random_state,
),
[0.001, 1, 1000],
),
(
"Infinite mixture with a Dirichlet process\n prior and" r"$\gamma_0=$",
BayesianGaussianMixture(
weight_concentration_prior_type="dirichlet_process",
n_components=2 * n_components,
reg_covar=0,
init_params="random",
max_iter=1500,
mean_precision_prior=0.8,
random_state=random_state,
),
[1, 1000, 100000],
),
]
# 生成数据
rng = np.random.RandomState(random_state)
X = np.vstack(
[
rng.multivariate_normal(means[j], covars[j], samples[j])
for j in range(n_components)
]
)
y = np.concatenate([np.full(samples[j], j, dtype=int) for j in range(n_components)])
# 在两个不同的图中绘制结果
for title, estimator, concentrations_prior in estimators:
plt.figure(figsize=(4.7 * 3, 8))
plt.subplots_adjust(
bottom=0.04, top=0.90, hspace=0.05, wspace=0.05, left=0.03, right=0.99
)
gs = gridspec.GridSpec(3, len(concentrations_prior))
for k, concentration in enumerate(concentrations_prior):
estimator.weight_concentration_prior = concentration
estimator.fit(X)
plot_results(
plt.subplot(gs[0:2, k]),
plt.subplot(gs[2, k]),
estimator,
X,
y,
r"%s$%.1e$" % (title, concentration),
plot_title=k == 0,
)
plt.show()
Total running time of the script: (0 minutes 2.875 seconds)
Related examples