Note
Go to the end to download the full example code. or to run this example in your browser via Binder
scikit-learn 1.4 版本发布亮点#
我们很高兴地宣布发布 scikit-learn 1.4 版本!此次更新包含了许多错误修复和改进,以及一些新的关键功能。以下是本次发布的一些主要功能。 有关所有更改的详尽列表 ,请参阅 release notes 。
要安装最新版本(使用 pip):
pip install --upgrade scikit-learn
或使用 conda:
conda install -c conda-forge scikit-learn
HistGradientBoosting 原生支持 DataFrame 中的分类数据类型#
ensemble.HistGradientBoostingClassifier 和 ensemble.HistGradientBoostingRegressor 现在直接支持包含分类特征的数据框。这里我们有一个包含分类和数值特征混合的数据集:
from sklearn.datasets import fetch_openml
X_adult, y_adult = fetch_openml("adult", version=2, return_X_y=True)
# 删除冗余和非特征列
X_adult = X_adult.drop(["education-num", "fnlwgt"], axis="columns")
X_adult.dtypes
age int64
workclass category
education category
marital-status category
occupation category
relationship category
race category
sex category
capital-gain int64
capital-loss int64
hours-per-week int64
native-country category
dtype: object
通过设置 categorical_features="from_dtype" ,梯度提升分类器在算法中将具有分类数据类型的列视为分类特征:
from sklearn.ensemble import HistGradientBoostingClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_auc_score
X_train, X_test, y_train, y_test = train_test_split(X_adult, y_adult, random_state=0)
hist = HistGradientBoostingClassifier(categorical_features="from_dtype")
hist.fit(X_train, y_train)
y_decision = hist.decision_function(X_test)
print(f"ROC AUC score is {roc_auc_score(y_test, y_decision)}")
ROC AUC score is 0.9283259911861934
Polars 输出在 set_output#
scikit-learn 的转换器现在通过 set_output API 支持 Polars 输出。
import polars as pl
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import ColumnTransformer
df = pl.DataFrame(
{"height": [120, 140, 150, 110, 100], "pet": ["dog", "cat", "dog", "cat", "cat"]}
)
preprocessor = ColumnTransformer(
[
("numerical", StandardScaler(), ["height"]),
("categorical", OneHotEncoder(sparse_output=False), ["pet"]),
],
verbose_feature_names_out=False,
)
preprocessor.set_output(transform="polars")
df_out = preprocessor.fit_transform(df)
df_out
print(f"Output type: {type(df_out)}")
Output type: <class 'polars.dataframe.frame.DataFrame'>
随机森林的缺失值支持#
类 ensemble.RandomForestClassifier 和 ensemble.RandomForestRegressor 现在支持缺失值。在训练每棵单独的树时,分割器会评估每个潜在的阈值,并将缺失值分别分配到左节点和右节点。更多细节请参见 用户指南 。
import numpy as np
from sklearn.ensemble import RandomForestClassifier
X = np.array([0, 1, 6, np.nan]).reshape(-1, 1)
y = [0, 0, 1, 1]
forest = RandomForestClassifier(random_state=0).fit(X, y)
forest.predict(X)
array([0, 0, 1, 1])
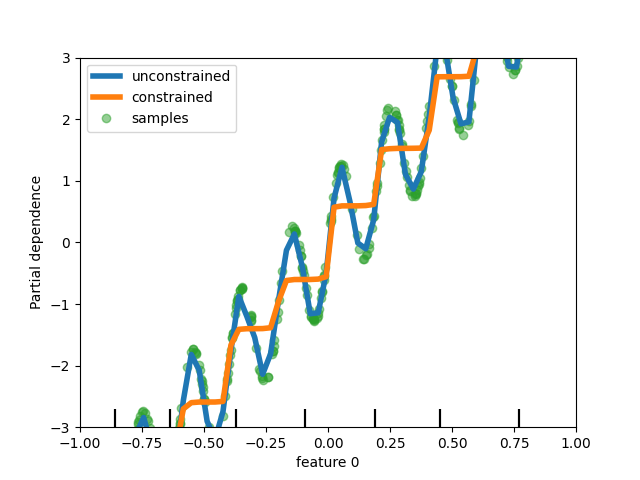
为树模型添加单调约束支持#
虽然我们在scikit-learn 0.23中为基于直方图的梯度提升添加了单调约束支持,但现在我们为所有其他树模型(如决策树、随机森林、极限树和精确梯度提升)提供了此功能。这里,我们在一个回归问题上展示了随机森林的这一特性。
import matplotlib.pyplot as plt
from sklearn.inspection import PartialDependenceDisplay
from sklearn.ensemble import RandomForestRegressor
n_samples = 500
rng = np.random.RandomState(0)
X = rng.randn(n_samples, 2)
noise = rng.normal(loc=0.0, scale=0.01, size=n_samples)
y = 5 * X[:, 0] + np.sin(10 * np.pi * X[:, 0]) - noise
rf_no_cst = RandomForestRegressor().fit(X, y)
rf_cst = RandomForestRegressor(monotonic_cst=[1, 0]).fit(X, y)
disp = PartialDependenceDisplay.from_estimator(
rf_no_cst,
X,
features=[0],
feature_names=["feature 0"],
line_kw={"linewidth": 4, "label": "unconstrained", "color": "tab:blue"},
)
PartialDependenceDisplay.from_estimator(
rf_cst,
X,
features=[0],
line_kw={"linewidth": 4, "label": "constrained", "color": "tab:orange"},
ax=disp.axes_,
)
disp.axes_[0, 0].plot(
X[:, 0], y, "o", alpha=0.5, zorder=-1, label="samples", color="tab:green"
)
disp.axes_[0, 0].set_ylim(-3, 3)
disp.axes_[0, 0].set_xlim(-1, 1)
disp.axes_[0, 0].legend()
plt.show()
增强的估计器显示#
估计器显示已被增强:如果我们查看上面定义的 forest :
forest
可以通过点击图表右上角的“?”图标来访问估计器的文档。
此外,当估计器拟合时,显示屏的颜色会从橙色变为蓝色。您也可以通过将鼠标悬停在“i”图标上获取此信息。
from sklearn.base import clone
clone(forest) # the clone is not fitted
元数据路由支持#
许多元估计器和交叉验证程序现在支持元数据路由,具体列表请参见:ref:用户指南<metadata_routing_models> 。例如,以下是使用样本权重和:class:~model_selection.GroupKFold 进行嵌套交叉验证的方法:
import sklearn
from sklearn.metrics import get_scorer
from sklearn.datasets import make_regression
from sklearn.linear_model import Lasso
from sklearn.model_selection import GridSearchCV, cross_validate, GroupKFold
# 目前,元数据路由默认是禁用的,需要显式启用。
sklearn.set_config(enable_metadata_routing=True)
n_samples = 100
X, y = make_regression(n_samples=n_samples, n_features=5, noise=0.5)
rng = np.random.RandomState(7)
groups = rng.randint(0, 10, size=n_samples)
sample_weights = rng.rand(n_samples)
estimator = Lasso().set_fit_request(sample_weight=True)
hyperparameter_grid = {"alpha": [0.1, 0.5, 1.0, 2.0]}
scoring_inner_cv = get_scorer("neg_mean_squared_error").set_score_request(
sample_weight=True
)
inner_cv = GroupKFold(n_splits=5)
grid_search = GridSearchCV(
estimator=estimator,
param_grid=hyperparameter_grid,
cv=inner_cv,
scoring=scoring_inner_cv,
)
outer_cv = GroupKFold(n_splits=5)
scorers = {
"mse": get_scorer("neg_mean_squared_error").set_score_request(sample_weight=True)
}
results = cross_validate(
grid_search,
X,
y,
cv=outer_cv,
scoring=scorers,
return_estimator=True,
params={"sample_weight": sample_weights, "groups": groups},
)
print("cv error on test sets:", results["test_mse"])
# 将标志设置为默认的 `False` 以避免与其他脚本发生干扰。
sklearn.set_config(enable_metadata_routing=False)
cv error on test sets: [-0.3429531 -0.36222461 -0.50291983 -0.32317095 -0.30104659]
改进了稀疏数据上PCA的内存和运行时效率#
现在,PCA能够本地处理 arpack 求解器的稀疏矩阵,通过利用 scipy.sparse.linalg.LinearOperator ,在执行数据集协方差矩阵的特征值分解时避免了大规模稀疏矩阵的具体化。
from sklearn.decomposition import PCA
import scipy.sparse as sp
from time import time
X_sparse = sp.random(m=1000, n=1000, random_state=0)
X_dense = X_sparse.toarray()
t0 = time()
PCA(n_components=10, svd_solver="arpack").fit(X_sparse)
time_sparse = time() - t0
t0 = time()
PCA(n_components=10, svd_solver="arpack").fit(X_dense)
time_dense = time() - t0
print(f"Speedup: {time_dense / time_sparse:.1f}x")
Speedup: 7.5x
Total running time of the script: (0 minutes 2.069 seconds)
Related examples