Note
Go to the end to download the full example code. or to run this example in your browser via Binder
模型复杂度影响#
演示模型复杂度如何影响预测准确性和计算性能。
- 我们将使用两个数据集:
Diabetes dataset 用于回归。 该数据集由糖尿病患者的10项测量值组成。 任务是预测疾病进展;
The 20 newsgroups text dataset 用于分类。该数据集由新闻组帖子组成。 任务是预测帖子所讨论的主题(20个主题中的一个)。
- 我们将对三种不同的估计器建模复杂度的影响:
SGDClassifier(用于分类数据), 实现随机梯度下降学习;NuSVR(用于回归数据), 实现Nu支持向量回归;GradientBoostingRegressor以前向阶段方式构建加性模型。 注意,从中等规模的数据集(n_samples >= 10_000)开始,HistGradientBoostingRegressor比GradientBoostingRegressor快得多, 但这不适用于本例。
我们通过选择每个模型中的相关参数来使模型复杂度变化。接下来,我们将测量对计算性能(延迟)和预测能力(MSE或Hamming Loss)的影响。
# 作者:scikit-learn 开发者
# SPDX 许可证标识符:BSD-3-Clause
import time
import matplotlib.pyplot as plt
import numpy as np
from sklearn import datasets
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.linear_model import SGDClassifier
from sklearn.metrics import hamming_loss, mean_squared_error
from sklearn.model_selection import train_test_split
from sklearn.svm import NuSVR
# 初始化随机生成器
np.random.seed(0)
加载数据#
首先我们加载两个数据集。
Note
我们使用 fetch_20newsgroups_vectorized 下载 20 个新闻组数据集。它返回可直接使用的特征。
Note
20类新闻组数据集的 X 是一个稀疏矩阵,而糖尿病数据集的 X 是一个numpy数组。
def generate_data(case):
"""生成回归/分类数据。"""
if case == "regression":
X, y = datasets.load_diabetes(return_X_y=True)
train_size = 0.8
elif case == "classification":
X, y = datasets.fetch_20newsgroups_vectorized(subset="all", return_X_y=True)
train_size = 0.4 # to make the example run faster
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=train_size, random_state=0
)
data = {"X_train": X_train, "X_test": X_test, "y_train": y_train, "y_test": y_test}
return data
regression_data = generate_data("regression")
classification_data = generate_data("classification")
基准影响#
接下来,我们可以计算参数对给定估计器的影响。在每一轮中,我们将使用新的 changing_param 值设置估计器,并收集预测时间、预测性能和复杂性,以观察这些变化如何影响估计器。我们将使用作为参数传递的 complexity_computer 来计算复杂性。
def benchmark_influence(conf):
"""
基准测试 `changing_param` 对MSE和延迟的影响。
"""
prediction_times = []
prediction_powers = []
complexities = []
for param_value in conf["changing_param_values"]:
conf["tuned_params"][conf["changing_param"]] = param_value
estimator = conf["estimator"](**conf["tuned_params"])
print("Benchmarking %s" % estimator)
estimator.fit(conf["data"]["X_train"], conf["data"]["y_train"])
conf["postfit_hook"](estimator)
complexity = conf["complexity_computer"](estimator)
complexities.append(complexity)
start_time = time.time()
for _ in range(conf["n_samples"]):
y_pred = estimator.predict(conf["data"]["X_test"])
elapsed_time = (time.time() - start_time) / float(conf["n_samples"])
prediction_times.append(elapsed_time)
pred_score = conf["prediction_performance_computer"](
conf["data"]["y_test"], y_pred
)
prediction_powers.append(pred_score)
print(
"Complexity: %d | %s: %.4f | Pred. Time: %fs\n"
% (
complexity,
conf["prediction_performance_label"],
pred_score,
elapsed_time,
)
)
return prediction_powers, prediction_times, complexities
选择参数#
我们通过创建一个包含所有必要值的字典来选择每个估计器的参数。
changing_param 是在每个估计器中变化的参数名称。
复杂度将由 complexity_label 定义,并使用 complexity_computer 计算。
还请注意,根据估计器类型的不同,我们传递的数据也不同。
def _count_nonzero_coefficients(estimator):
a = estimator.coef_.toarray()
return np.count_nonzero(a)
configurations = [
{
"estimator": SGDClassifier,
"tuned_params": {
"penalty": "elasticnet",
"alpha": 0.001,
"loss": "modified_huber",
"fit_intercept": True,
"tol": 1e-1,
"n_iter_no_change": 2,
},
"changing_param": "l1_ratio",
"changing_param_values": [0.25, 0.5, 0.75, 0.9],
"complexity_label": "non_zero coefficients",
"complexity_computer": _count_nonzero_coefficients,
"prediction_performance_computer": hamming_loss,
"prediction_performance_label": "Hamming Loss (Misclassification Ratio)",
"postfit_hook": lambda x: x.sparsify(),
"data": classification_data,
"n_samples": 5,
},
{
"estimator": NuSVR,
"tuned_params": {"C": 1e3, "gamma": 2**-15},
"changing_param": "nu",
"changing_param_values": [0.05, 0.1, 0.2, 0.35, 0.5],
"complexity_label": "n_support_vectors",
"complexity_computer": lambda x: len(x.support_vectors_),
"data": regression_data,
"postfit_hook": lambda x: x,
"prediction_performance_computer": mean_squared_error,
"prediction_performance_label": "MSE",
"n_samples": 15,
},
{
"estimator": GradientBoostingRegressor,
"tuned_params": {
"loss": "squared_error",
"learning_rate": 0.05,
"max_depth": 2,
},
"changing_param": "n_estimators",
"changing_param_values": [10, 25, 50, 75, 100],
"complexity_label": "n_trees",
"complexity_computer": lambda x: x.n_estimators,
"data": regression_data,
"postfit_hook": lambda x: x,
"prediction_performance_computer": mean_squared_error,
"prediction_performance_label": "MSE",
"n_samples": 15,
},
]
运行代码并绘制结果
我们定义了运行基准测试所需的所有函数。现在,我们将循环遍历之前定义的不同配置。随后,我们可以分析从基准测试中获得的图表:
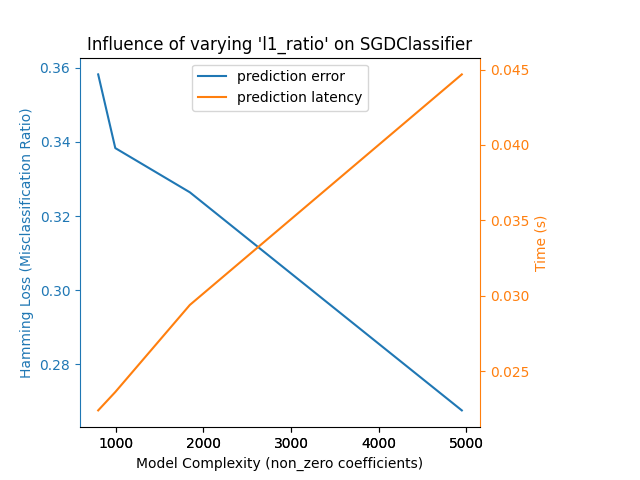
放松SGD分类器中的 L1 惩罚会减少预测误差,但会增加训练时间。
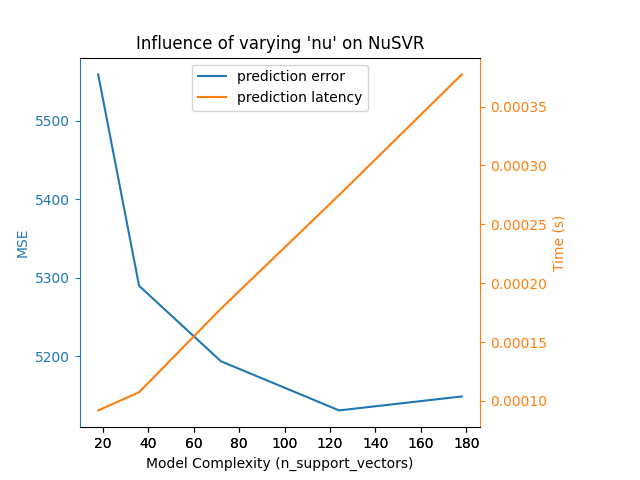
我们可以对Nu-SVR的训练时间进行类似的分析,训练时间随着支持向量的数量增加而增加。然而,我们观察到存在一个最佳的支持向量数量,可以减少预测误差。确实,支持向量太少会导致模型欠拟合,而支持向量太多会导致模型过拟合。
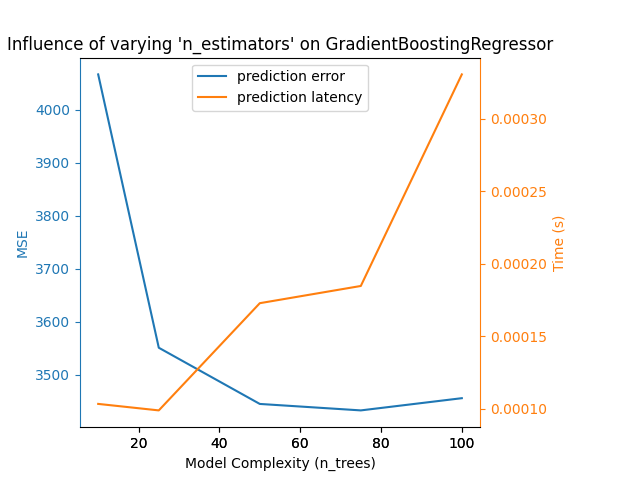
对于梯度提升模型也可以得出完全相同的结论。与Nu-SVR的唯一区别是,集成中树的数量过多并不会造成同样的负面影响。
def plot_influence(conf, mse_values, prediction_times, complexities):
"""
绘制模型复杂度对准确性和延迟的影响。
"""
fig = plt.figure()
fig.subplots_adjust(right=0.75)
# 第一轴(预测误差)
ax1 = fig.add_subplot(111)
line1 = ax1.plot(complexities, mse_values, c="tab:blue", ls="-")[0]
ax1.set_xlabel("Model Complexity (%s)" % conf["complexity_label"])
y1_label = conf["prediction_performance_label"]
ax1.set_ylabel(y1_label)
ax1.spines["left"].set_color(line1.get_color())
ax1.yaxis.label.set_color(line1.get_color())
ax1.tick_params(axis="y", colors=line1.get_color())
# 第二轴(延迟)
ax2 = fig.add_subplot(111, sharex=ax1, frameon=False)
line2 = ax2.plot(complexities, prediction_times, c="tab:orange", ls="-")[0]
ax2.yaxis.tick_right()
ax2.yaxis.set_label_position("right")
y2_label = "Time (s)"
ax2.set_ylabel(y2_label)
ax1.spines["right"].set_color(line2.get_color())
ax2.yaxis.label.set_color(line2.get_color())
ax2.tick_params(axis="y", colors=line2.get_color())
plt.legend(
(line1, line2), ("prediction error", "prediction latency"), loc="upper center"
)
plt.title(
"Influence of varying '%s' on %s"
% (conf["changing_param"], conf["estimator"].__name__)
)
for conf in configurations:
prediction_performances, prediction_times, complexities = benchmark_influence(conf)
plot_influence(conf, prediction_performances, prediction_times, complexities)
plt.show()
Benchmarking SGDClassifier(alpha=0.001, l1_ratio=0.25, loss='modified_huber',
n_iter_no_change=2, penalty='elasticnet', tol=0.1)
Complexity: 4948 | Hamming Loss (Misclassification Ratio): 0.2675 | Pred. Time: 0.044682s
Benchmarking SGDClassifier(alpha=0.001, l1_ratio=0.5, loss='modified_huber',
n_iter_no_change=2, penalty='elasticnet', tol=0.1)
Complexity: 1847 | Hamming Loss (Misclassification Ratio): 0.3264 | Pred. Time: 0.029387s
Benchmarking SGDClassifier(alpha=0.001, l1_ratio=0.75, loss='modified_huber',
n_iter_no_change=2, penalty='elasticnet', tol=0.1)
Complexity: 997 | Hamming Loss (Misclassification Ratio): 0.3383 | Pred. Time: 0.023611s
Benchmarking SGDClassifier(alpha=0.001, l1_ratio=0.9, loss='modified_huber',
n_iter_no_change=2, penalty='elasticnet', tol=0.1)
Complexity: 802 | Hamming Loss (Misclassification Ratio): 0.3582 | Pred. Time: 0.022392s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05, nu=0.05)
Complexity: 18 | MSE: 5558.7313 | Pred. Time: 0.000092s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05, nu=0.1)
Complexity: 36 | MSE: 5289.8022 | Pred. Time: 0.000107s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05, nu=0.2)
Complexity: 72 | MSE: 5193.8353 | Pred. Time: 0.000178s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05, nu=0.35)
Complexity: 124 | MSE: 5131.3279 | Pred. Time: 0.000275s
Benchmarking NuSVR(C=1000.0, gamma=3.0517578125e-05)
Complexity: 178 | MSE: 5149.0779 | Pred. Time: 0.000377s
Benchmarking GradientBoostingRegressor(learning_rate=0.05, max_depth=2, n_estimators=10)
Complexity: 10 | MSE: 4066.4812 | Pred. Time: 0.000103s
Benchmarking GradientBoostingRegressor(learning_rate=0.05, max_depth=2, n_estimators=25)
Complexity: 25 | MSE: 3551.1723 | Pred. Time: 0.000099s
Benchmarking GradientBoostingRegressor(learning_rate=0.05, max_depth=2, n_estimators=50)
Complexity: 50 | MSE: 3445.2171 | Pred. Time: 0.000173s
Benchmarking GradientBoostingRegressor(learning_rate=0.05, max_depth=2, n_estimators=75)
Complexity: 75 | MSE: 3433.0358 | Pred. Time: 0.000185s
Benchmarking GradientBoostingRegressor(learning_rate=0.05, max_depth=2)
Complexity: 100 | MSE: 3456.0602 | Pred. Time: 0.000331s
Conclusion#
作为结论,我们可以得出以下见解:
一个更复杂(或更具表现力)的模型将需要更长的训练时间;
一个更复杂的模型并不保证能减少预测误差。
这些方面与模型的泛化能力以及避免模型欠拟合或过拟合有关。
Total running time of the script: (0 minutes 8.516 seconds)
Related examples