Note
Go to the end to download the full example code. or to run this example in your browser via Binder
使用概率PCA和因子分析(FA)进行模型选择#
概率PCA和因子分析是概率模型。 其结果是可以使用新数据的似然性进行模型选择和协方差估计。 在这里,我们通过交叉验证比较了在低秩数据上添加同方差噪声(噪声方差 对于每个特征都是相同的)或异方差噪声(噪声方差对于每个特征是不同的)时的PCA和FA。 在第二步中,我们将模型的似然性与从收缩协方差估计器获得的似然性进行比较。
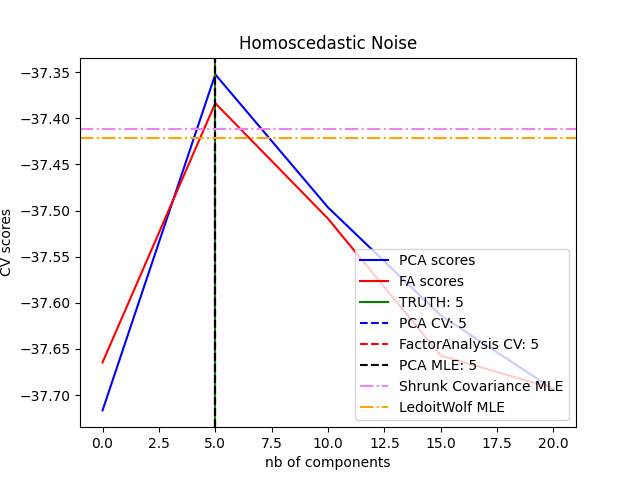
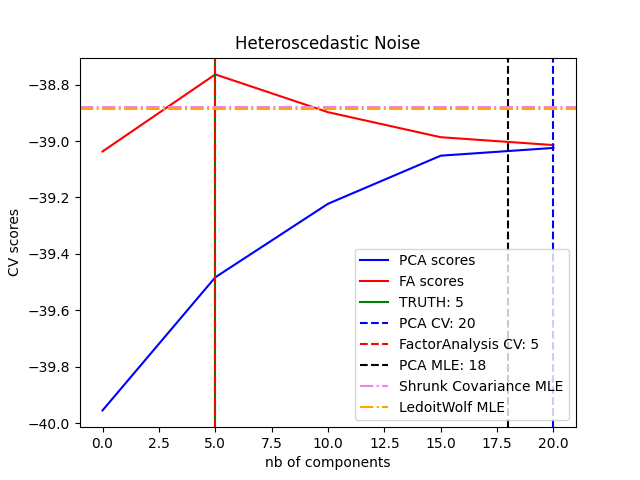
可以观察到,在同方差噪声下,FA和PCA都成功地恢复了低秩子空间的大小。 在这种情况下,PCA的似然性高于FA。然而,当存在异方差噪声时,PCA失败并高估了秩。 在适当的情况下(选择组件的数量),低秩模型的保留数据比收缩模型更有可能。
还比较了Thomas P. Minka在《PCA维度的自动选择》中提出的自动估计方法。 NIPS 2000: 598-604
# 作者:scikit-learn 开发者
# SPDX-License-Identifier: BSD-3-Clause
创建数据#
import numpy as np
from scipy import linalg
n_samples, n_features, rank = 500, 25, 5
sigma = 1.0
rng = np.random.RandomState(42)
U, _, _ = linalg.svd(rng.randn(n_features, n_features))
X = np.dot(rng.randn(n_samples, rank), U[:, :rank].T)
# 添加同方差噪声
X_homo = X + sigma * rng.randn(n_samples, n_features)
# 添加异方差噪声
sigmas = sigma * rng.rand(n_features) + sigma / 2.0
X_hetero = X + rng.randn(n_samples, n_features) * sigmas
拟合模型#
import matplotlib.pyplot as plt
from sklearn.covariance import LedoitWolf, ShrunkCovariance
from sklearn.decomposition import PCA, FactorAnalysis
from sklearn.model_selection import GridSearchCV, cross_val_score
n_components = np.arange(0, n_features, 5) # options for n_components
def compute_scores(X):
pca = PCA(svd_solver="full")
fa = FactorAnalysis()
pca_scores, fa_scores = [], []
for n in n_components:
pca.n_components = n
fa.n_components = n
pca_scores.append(np.mean(cross_val_score(pca, X)))
fa_scores.append(np.mean(cross_val_score(fa, X)))
return pca_scores, fa_scores
def shrunk_cov_score(X):
shrinkages = np.logspace(-2, 0, 30)
cv = GridSearchCV(ShrunkCovariance(), {"shrinkage": shrinkages})
return np.mean(cross_val_score(cv.fit(X).best_estimator_, X))
def lw_score(X):
return np.mean(cross_val_score(LedoitWolf(), X))
for X, title in [(X_homo, "Homoscedastic Noise"), (X_hetero, "Heteroscedastic Noise")]:
pca_scores, fa_scores = compute_scores(X)
n_components_pca = n_components[np.argmax(pca_scores)]
n_components_fa = n_components[np.argmax(fa_scores)]
pca = PCA(svd_solver="full", n_components="mle")
pca.fit(X)
n_components_pca_mle = pca.n_components_
print("best n_components by PCA CV = %d" % n_components_pca)
print("best n_components by FactorAnalysis CV = %d" % n_components_fa)
print("best n_components by PCA MLE = %d" % n_components_pca_mle)
plt.figure()
plt.plot(n_components, pca_scores, "b", label="PCA scores")
plt.plot(n_components, fa_scores, "r", label="FA scores")
plt.axvline(rank, color="g", label="TRUTH: %d" % rank, linestyle="-")
plt.axvline(
n_components_pca,
color="b",
label="PCA CV: %d" % n_components_pca,
linestyle="--",
)
plt.axvline(
n_components_fa,
color="r",
label="FactorAnalysis CV: %d" % n_components_fa,
linestyle="--",
)
plt.axvline(
n_components_pca_mle,
color="k",
label="PCA MLE: %d" % n_components_pca_mle,
linestyle="--",
)
# 与其他协方差估计器进行比较
plt.axhline(
shrunk_cov_score(X),
color="violet",
label="Shrunk Covariance MLE",
linestyle="-.",
)
plt.axhline(
lw_score(X),
color="orange",
label="LedoitWolf MLE" % n_components_pca_mle,
linestyle="-.",
)
plt.xlabel("nb of components")
plt.ylabel("CV scores")
plt.legend(loc="lower right")
plt.title(title)
plt.show()
best n_components by PCA CV = 5
best n_components by FactorAnalysis CV = 5
best n_components by PCA MLE = 5
best n_components by PCA CV = 20
best n_components by FactorAnalysis CV = 5
best n_components by PCA MLE = 18
Total running time of the script: (0 minutes 1.812 seconds)
Related examples