Note
Go to the end to download the full example code. or to run this example in your browser via Binder
分类器的概率校准#
在进行分类时,你通常不仅希望预测类别标签,还希望预测相关的概率。这个概率给你对预测的一种置信度。然而,并不是所有的分类器都能提供良好校准的概率,有些分类器过于自信,而有些则信心不足。因此,通常需要对预测的概率进行单独的后处理校准。这个例子展示了两种不同的校准方法,并使用Brier得分(参见https://en.wikipedia.org/wiki/Brier_score)评估返回概率的质量。
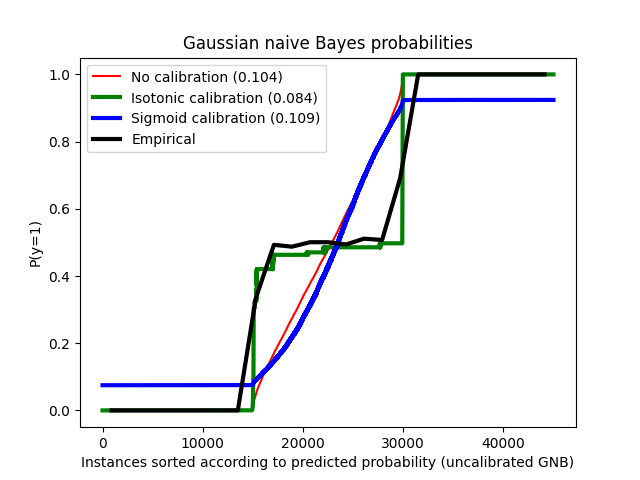
比较了使用高斯朴素贝叶斯分类器在没有校准、使用Sigmoid校准和使用非参数的Isotonic校准下的估计概率。可以观察到,只有非参数模型能够提供概率校准,使得大多数属于中间簇且标签异质的样本的概率接近预期的0.5。这显著改善了Brier得分。
# 作者:scikit-learn 开发者
# SPDX 许可证标识符:BSD-3-Clause
生成合成数据集#
import numpy as np
from sklearn.datasets import make_blobs
from sklearn.model_selection import train_test_split
n_samples = 50000
n_bins = 3 # use 3 bins for calibration_curve as we have 3 clusters here
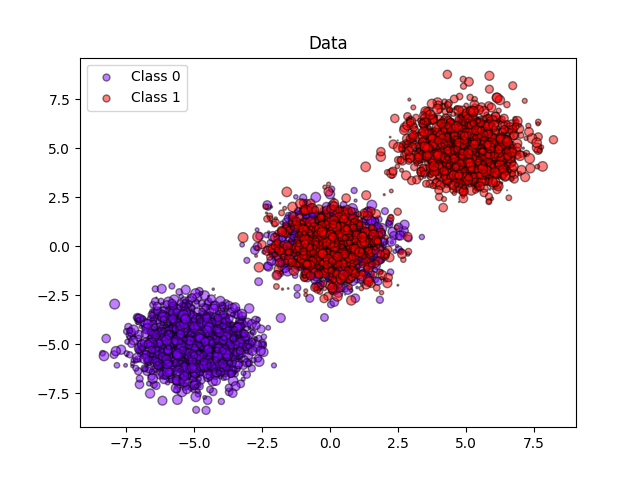
# 生成3个数据簇,其中包含2个类别,其中第二个数据簇包含一半正样本和一半负样本。因此,该数据簇中的概率为0.5。
centers = [(-5, -5), (0, 0), (5, 5)]
X, y = make_blobs(n_samples=n_samples, centers=centers, shuffle=False, random_state=42)
y[: n_samples // 2] = 0
y[n_samples // 2 :] = 1
sample_weight = np.random.RandomState(42).rand(y.shape[0])
# 划分训练集和测试集以进行校准
X_train, X_test, y_train, y_test, sw_train, sw_test = train_test_split(
X, y, sample_weight, test_size=0.9, random_state=42
)
高斯朴素贝叶斯#
from sklearn.calibration import CalibratedClassifierCV
from sklearn.metrics import brier_score_loss
from sklearn.naive_bayes import GaussianNB
# 无需校准
clf = GaussianNB()
clf.fit(X_train, y_train) # GaussianNB itself does not support sample-weights
prob_pos_clf = clf.predict_proba(X_test)[:, 1]
# 通过等渗校准
clf_isotonic = CalibratedClassifierCV(clf, cv=2, method="isotonic")
clf_isotonic.fit(X_train, y_train, sample_weight=sw_train)
prob_pos_isotonic = clf_isotonic.predict_proba(X_test)[:, 1]
# 使用Sigmoid校准
clf_sigmoid = CalibratedClassifierCV(clf, cv=2, method="sigmoid")
clf_sigmoid.fit(X_train, y_train, sample_weight=sw_train)
prob_pos_sigmoid = clf_sigmoid.predict_proba(X_test)[:, 1]
print("Brier score losses: (the smaller the better)")
clf_score = brier_score_loss(y_test, prob_pos_clf, sample_weight=sw_test)
print("No calibration: %1.3f" % clf_score)
clf_isotonic_score = brier_score_loss(y_test, prob_pos_isotonic, sample_weight=sw_test)
print("With isotonic calibration: %1.3f" % clf_isotonic_score)
clf_sigmoid_score = brier_score_loss(y_test, prob_pos_sigmoid, sample_weight=sw_test)
print("With sigmoid calibration: %1.3f" % clf_sigmoid_score)
Brier score losses: (the smaller the better)
No calibration: 0.104
With isotonic calibration: 0.084
With sigmoid calibration: 0.109
绘制数据和预测概率#
import matplotlib.pyplot as plt
from matplotlib import cm
plt.figure()
y_unique = np.unique(y)
colors = cm.rainbow(np.linspace(0.0, 1.0, y_unique.size))
for this_y, color in zip(y_unique, colors):
this_X = X_train[y_train == this_y]
this_sw = sw_train[y_train == this_y]
plt.scatter(
this_X[:, 0],
this_X[:, 1],
s=this_sw * 50,
c=color[np.newaxis, :],
alpha=0.5,
edgecolor="k",
label="Class %s" % this_y,
)
plt.legend(loc="best")
plt.title("Data")
plt.figure()
order = np.lexsort((prob_pos_clf,))
plt.plot(prob_pos_clf[order], "r", label="No calibration (%1.3f)" % clf_score)
plt.plot(
prob_pos_isotonic[order],
"g",
linewidth=3,
label="Isotonic calibration (%1.3f)" % clf_isotonic_score,
)
plt.plot(
prob_pos_sigmoid[order],
"b",
linewidth=3,
label="Sigmoid calibration (%1.3f)" % clf_sigmoid_score,
)
plt.plot(
np.linspace(0, y_test.size, 51)[1::2],
y_test[order].reshape(25, -1).mean(1),
"k",
linewidth=3,
label=r"Empirical",
)
plt.ylim([-0.05, 1.05])
plt.xlabel("Instances sorted according to predicted probability (uncalibrated GNB)")
plt.ylabel("P(y=1)")
plt.legend(loc="upper left")
plt.title("Gaussian naive Bayes probabilities")
plt.show()
Total running time of the script: (0 minutes 0.170 seconds)
Related examples